1.3: DNA Mutations
- Page ID
- 73668
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- List the types of mutation.
- Predict the effects of specific mutations in gene coding regions on the protein encoded by the gene.
- Describe ways in which mutations arise, naturally and experimentally.
Introduction
Crop improvement relies on genetic variation, which is synonymous with DNA variation. Mutations are the source of genetic variation that is the driver for trait improvement. In general, mutations are rare events, and most are deleterious and get selected against during the course of evolution. However, occasionally mutations create genetic variation that is beneficial. As such, it is often of interest for plant improvement or for genetic studies to induce genetic mutations by exposing seed or plant tissue to certain agents, called mutagens.
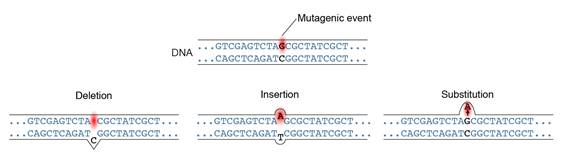
By definition, a mutation is a heritable change in DNA sequence. This can happen in several ways: substitution of a DNA base, insertion or deletion of one or more DNA bases, or by large-scale chromosomal rearrangements, the latter of which will not be considered here. It has also become clear in recent years that chromatin carries additional information to the DNA sequence in the form of base modifications such as cytosine methylation. This is known as epigenetic modification and can impact gene function and plant traits.
A. DNA base substitutions
The simplest type of mutation is a substitution of one base for another in the DNA sequence. Substitutions most often arise as errors during DNA replication or repair. The most common type is the transition, where one pyrimidine may be substituted by the other, or one purine by the other. The less common type is the transversion, in which a purine may be substituted by a pyrimidine or vice versa. As described in the next section, these can have various effects on gene function depending on where they occur in a gene.
B. Insertions & deletions

Insertions or deletions of DNA are other types of mutations that occur quite frequently. They can vary in size from one to thousands or more nucleotide bases. Their effects on gene function depend on the size of the insertion or deletion, and the location relative to the gene. As described below, insertions and deletions that are in multiples of 3 are referred to as in-frame insertions and deletions and result in the addition or deletion of amino acids from the protein sequence. When not in multiples of three, insertions and deletions lead to frameshift mutations, altering the amino acid sequence of the protein encoded by the mutated gene.

A common source of inserted DNA is from transposons, sequences of DNA that are able to move from one genomic site and insert into another. Transposons were discovered in maize by Barbara McClintock. For this discovery, she was awarded the Nobel Prize in Physiology and Medicine in 1983.
Some transposons copy themselves and multiply in number within a genome. Transposons can cause wide scale chromosome rearrangements and gene mutations. A feature of mutations caused by many plant transposons is that they are unstable. Excision of a transposon from the mutated gene can often restore it to wild type or create a new stable mutant allele. Transposons are found in every living organism studied and are a major driving force in genome evolution.

Effects of mutations on gene function
A. Coding region mutations
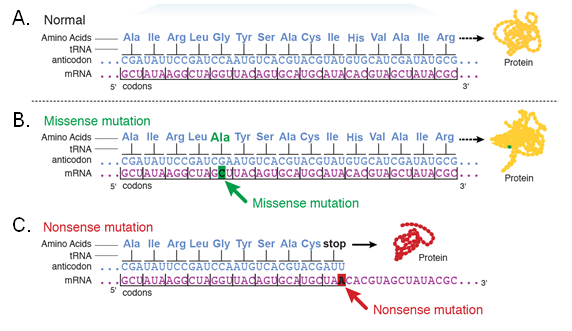
Silent mutation
A silent mutation is a mutation that results in the change of a codon without a change in the amino acid represented by the codon. Because of the redundancy of the genetic code, DNA base substitutions may not lead to the incorporation of an incorrect amino acid in the protein.
Missense mutation (amino acid substitution)
A missense mutation is a single nucleotide base substitution that alters a codon such that it codes for a different amino acid that is incorporated into the encoded protein. The amino acid substitution may affect the function of the protein if it occurs in a critical portion of the protein.
Frameshift mutation
Due to the triplet nature of the genetic code, an insertion or deletion can change the reading frame for the entire subsequent sequence. For example, if a particular sequence is read sequentially (Figure 4).
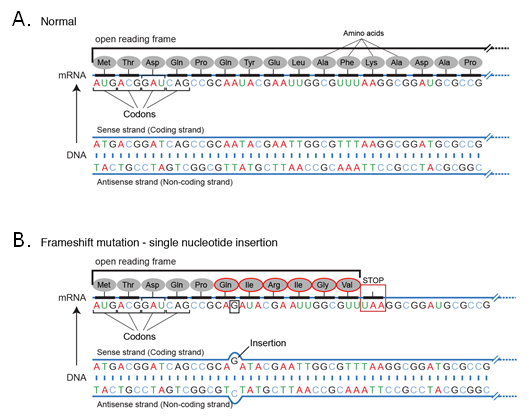
Because the new sequence of codons is different from the original, the entire amino acid sequence is changed from the point of insertion. A similar effect is seen for deletions. This effect is seen whenever the number of nucleotide bases inserted or deleted is not a multiple of 3 and it usually will result in the loss of the function of the protein.
Amino acid additions or deletions
On the other hand, if insertions or deletions are in multiples of 3, amino acid additions or deletions can arise. Consider the following 3-base insertion (XXX):
UUU CCC XXX AAA GGG (codons)
aa1 aa2 aa5 aa3 aa4 (amino acids)
If such insertions or deletions occur in important regions of a protein, they might impair function.
Nonsense mutation
A nonsense mutation arises when a functional codon is changed to a stop codon. Nonsense mutations may cause premature termination of translation.
B. Regulatory region mutations

Promoters and enhancers control temporal, spatial and quantitative aspects of gene expression. As such, mutations in these regulatory regions of a gene can alter the regulation of the gene’s expression. Genes can become expressed at inappropriate times, places or levels, which can affect a plant’s phenotype.
Teosinte, the progenitor of modern maize, produces multiple branches (Figure 6A) due to reduced apical dominance. In teosinte and modern maize, apical dominance is under the control of a gene called teosinte branched1, tb1 (Doebley et al. 1997). An insertion of a transposon in the regulatory region of the tb1 gene in modern maize causes the gene to be strongly expressed (Doebley et al. 1997; Studer et al. 2011), resulting in enhanced apical dominance, and suppression of excessive branching (Figure 6B).
C. Recessive vs. dominant mutations
Recessive mutations
Recessive mutations most commonly result from a loss of function. A loss of function mutation causes a decrease in the function of a gene. A complete loss of function is known as a null mutation while partial loss of function mutants are often referred to as hypomorphs or “leaky”. Loss of function mutations can affect gene expression or the function of the resultant protein. Most loss of function mutations are recessive because in a heterozygote, the normal allele can provide the necessary function.
Dominant mutations
Dominant mutations can affect genes in several different ways. One general class is gain of function mutations. These encompass several different types.
- Overexpression mutants. As the name implies, the affected gene is expressed at levels that are inappropriately high, causing too much of a gene product to be produced.
- Neomorphic mutations arise when a mutation causes a qualitatively new effect not seen with normal alleles. This can occur when a mutation affects the regulation of a gene such that it becomes expressed in a new time or place in the plant. It can also happen when a mutation alters the properties of the encoded protein. For example, a transcription factor could be mutated to recognize a different promoter sequence, or an enzyme could recognize a new substrate
- Dominant negative mutations alter a gene product such that the mutant gene product interferes with the function of the normal one. This can occur at the protein level. For example, a protein might function as a multimeric complex and the presence of one mutant subunit could compromise the function of the entire complex. Dominant negatives can also occur at the RNA level for example if an insertion, deletion or rearrangement causes the wrong strand to be transcribed, creating an antisense or RNAi suppression effect.
Effects of mutations on traits
Example of a loss of function mutation – gene function is required for a trait
Sweetness is an important quality trait in maize (corn). One of the loss of function mutants used to improve sweetness in corn is sugary1 (su1). The Su1 gene is expressed in the endosperm (Lertrat and Pulam, 2007), and it encodes and enzyme that is required for normal starch biosynthesis (Rahman et al. 1998). In the su1 mutant, with defective starch biosynthesis, the concentration of sugar is 3 times higher than wild type making this mutant valuable for sweet corn (Lertrat and Pulam, 2007). In addition, su1 kernels accumulate a highly branched polysaccharide called phytoglycogen, which gives the kernels a creamy texture. New commercial sweet corn hybrids are developed using a combination of starch biosynthesis mutants (Nelson and Pan, 1995; Lertrat and Pulam, 2007).
Example of a gain of function mutation – gene function is sufficient for a trait
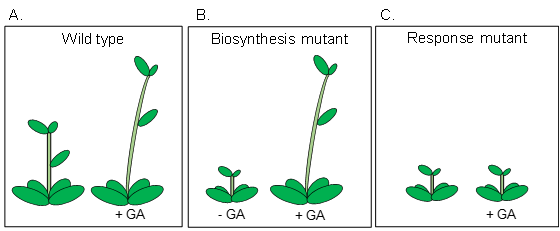
The plant growth hormone, gibberellin (GA) regulates plant height, thus, a plant that fails to produce GA, or does not respond to GA will have a dwarfed phenotype (Figure 7). At the molecular level, the cell response to GA involves several processes, including the destruction of repressor proteins, referred to as DELLAs.
The “green revolution gene” Reduced height-1 (Rht-1) from wheat encodes a DELLA repressor of GA signaling (Peng et al. 1999). A stretch of five amino acids referred to as the DELLA domain is required for the destruction of the repressor in response to GA. Mutations in the DELLA domain of Rht-1 genes result in a protein product that is resistant to degradation, leading to failure to signal a GA response. The failure to signal the presence of GA in the rht-1 mutants is responsible for their short stature. When the mutant rht-1 gene is transformed into rice, the result is also short plants, which is proof that the new function of the DELLA protein is sufficient to induce dwarfism (Peng et al. 1999).
Generation of mutations
Because mutations are the ultimate source of genetic variation for breeding and genetic studies, it is sometimes of interest to generate new mutations. New DNA mutations (Figure 8) can arise spontaneously or they can be induced by experimental methods.
Spontaneous mutations arise “naturally”
That is, no particular effort was made on the part of the scientist to increase the mutation rate. Spontaneous mutations generally occur at very low rates, on the order of 1 per gene per million gametes.
Mistakes in DNA replication
Very rarely, incorrect bases are incorporated or bases omitted from a DNA strand during synthesis. Such mistakes can lead to spontaneous substitutions, insertions or deletions. For example, strand slippage due to the formation of a loop.
Environmental mutagens
There are several chemical agents present in the environment with potential to damage DNA and create mutations. These include naturally occurring compounds as well as man-made pollutants. Some modify DNA nucleotide bases through deamination (e.g., nitrous acid), while others promote oxidative reactions that may damage DNA (e.g., ozone).
Radiation is another type of environmental mutagen. Ionizing radiation (e.g., X-rays) can shatter DNA sequences and promote chromosome rearrangements. The less powerful ultraviolet rays can penetrate the cell and promote the formation of pyrimidine dimers which may inhibit replication and transcription.
Experimental mutagenesis involves applying a mutagenic agent to a plant or plant part to generate a mutation. For the mutation to be useful, it must be heritable, which imposes limitations on what tissues can be treated. Following the mutagenic treatment, a subsequent breeding scheme must be implemented to generate pure breeding lines of appropriate genotypes to display mutant phenotypes. These will depend on the species and the design of the mutagenesis.
Chemical mutagenesis
A commonly used chemical mutagen for experimental plant biology and mutation breeding is ethyl methanesulfonate (EMS). The compound is known as an alkylating agent and induces a CG to TA transition.
Seed Mutagenesis
This is the most common method of chemical mutagenesis in most species. Seeds are soaked in EMS solution, planted and allowed to flower and set seed. During EMS mutagenesis, every cell in the embryo will be independently mutagenized and the resultant seed and subsequent plant will be chimeric. These seeds can be sown and plants screened for traits of interest. For self-pollinating species, the required breeding is very simple, whereas for dioecious species it would be much more elaborate and labor-intensive.
Pollen Mutagenesis
For a few species such as maize, pollen mutagenesis is the method of choice. Pollen are treated with EMS and applied to silks of an acceptor plant. The benefit is that the resultant seed is not chimeric but rather all the cells of the embryo carry the same mutagenized paternal genome inherited from the pollen grain.
Insertional mutagenesis
The idea behind insertional mutagenesis is that a mobile or introduced piece of DNA can sometimes insert into a gene, thereby inactivating or modifying the function of the gene. The approach is useful in gene cloning because the inserted DNA serves as a “tag” that can be used to locate the gene of interest, and subsequently isolate that gene.
Another strategy used by researchers is mutagenesis using T-DNA from the Agrobacterium tumefaciens Ti plasmid. A larger number of plants (up to 10,000 for Arabidopsis) are transformed with T-DNA to generate mutants at random.
Similar to the example of transposon mutagenesis above, a gene can be “tagged” by a T-DNA and the tag used to isolate the gene. However, a T-DNA insertions are stable and thus disrupt the gene permanently.
In recent years, insertional mutagens have been engineered in many innovative ways. For example, strong promoters have been engineered into transposons such that if the transposon inserts near a gene, it can cause strong transcriptional expression of that gene. Such “activation tagging” approaches have been successful in identifying many genes that were not amenable to other mutational approaches.
Mutations alter A-T and G-C base pairs in DNA. A mutation in a coding sequence may alter the sequence and function of the protein product. A frameshift mutation changes the reading frame through insertions or deletions to produce an entirely novel product. A point mutation on the other hand alters only the amino acid represented by the codon in which the mutation exists. Mutations in regulatory regions might alter the expression of a gene. Mutations have been powerful tools for genetic studies as well as to study biological functions, for example, growth and development. The natural occurrence of mutations can be enhanced experimentally by applying agents referred to as mutagens. Researchers have used certain mutagens to induce mutations to study gene function and apply new traits for crop improvement.
Activity 1
For this activity, it may be helpful to refer to the genetic code chart.
For the following DNA sequence:
5′ ATG TTG GAG AAG GTT GAA ACT TTC 3′
Write the coding strand
3′ TAC AAG CTC TTC CAA CTT TGA AAG 5′
Write the mRNA sequence from the given DNA sequence
AUG UUC GAG AAG GUU GAA ACU UUG
Write the resulting peptide sequence
Met-Phe-Glu-Lys-Val-Glu-Thr-Leu
If the third triplet of the original DNA sequence is mutated to GAA to result in the following sequence
5′ ATG TTG GAA AAG GTT GAA ACT TTC 3′
What will be the mRNA sequence from the mutated DNA sequence?
AUG UUC GAA AAG GUU GAA ACU UUG
What will be the peptide sequence from the mutated DNA sequence?
Met-Phe-Glu-Lys-Val-Glu-Thr-Leu
What type of coding region mutation is this? Give a reason for your answer.
- Silent
- Amino acid substitution
- Frameshift
- Nonsense
If the sixth triplet of the original DNA sequence is mutated to TAA to result in the following sequence
5′ ATG TTG GAG AAG GTT TAA ACT TTC 3′
What will be the mRNA sequence from the mutated DNA sequence?
AUG UUC GAG AAG GUU UAA ACU UUG
What will be the peptide sequence from the mutated DNA sequence?
Met-Phe-Glu-Lys-Val
What type of coding region mutation is this? Give reason for your answer.
- Silent
- Amino acid substitution
- Frameshift
- Nonsense
The original DNA sequence is mutated to result in the following sequence
5′ ATG TTG GAG AAG GTT CGA AAC TTT C 3′
What will be the mRNA sequence from the mutated DNA sequence?
AUG UUC GAG AAG GUU CGA AAC UUU C
What will be the peptide sequence from the mutated DNA sequence?
Met-Phe-Glu-Lys-Val-Arg-Asn-Phe
What type of coding region mutation is this? Give a reason for your answer.
- Silent
- Amino acid substitution
- Frameshift
- Nonsense
1. A hypothetical plant gene encodes a protein with the following amino acid sequence:
Met-Leu-Lys-Thr-Phe-Val-Glu
A mutation of a single nucleotide alters the amino acid sequence to:
Met-Tyr-Asp-Pro-Gly-Ala-Lys-Ile-Arg
Describe the type of the mutation and the position of the codon that is affected.
2. For the following DNA sequence
3′ ATG GCC GGC AAT CAA CTA TAT TGA 5′
1 24 (nucleotide number)
a. Write the coding strand
b. Write the mRNA strand
c. Write the protein strand
d. Give the altered amino acid sequence of the protein for each of the following mutations:
i. A transition at nucleotide 11
ii. A transition at nucleotide 13
iii. A single nucleotide deletion at nucleotide 7
iv. A T to A change at nucleotide 15
v. An addition of ACC after nucleotide 6
vi. A transition at nucleotide 9
References
Doebley et al. 1997. The evolution of apical dominance in maize. Nature 386: 485-488.
Lertrat, K., Pulam, T. 2007. Breeding for increased sweetness in sweet corn. Int. J. Plant. Breed. 1: 27-30.
Nelson, O., Pan, D. 1995. Starch synthesis in maize endosperms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46: 475-496.
Peng, J., Richards, D.E., Hartley, N.M., Murphy, G.P., Devos, K.M., Flintman, J.E., Beales, J., Fish, L.J., Worland, A.J., Pelica, F., Sudhakar, D., Christou, P., Snape, J.W., Gale, M.D., Harberd, N.P. 1999. “Green revolution” genes encode mutant gibberellin response modulators. Nature 400: 256-261.


