1.2: DNA- The Genetic Material
- Page ID
- 73667
- Detail the molecular nature of a gene.
- Recognize when and where replication of the DNA that is packaged in an organism’s chromosome is happening for growth, maintenance, and reproduction.
- Describe how chromosome replication speed can be increased with multiple origins of replication and bidirectional replication.
- Explain the role of each of the primary DNA replicating enzymes in conducting replication of a chromosome. Include the role of these enzyme in ensuring the accuracy of the replication process.
- Predict how the semiconservative replication of a double stranded DNA will happen given the structure of the molecule and the specific function of the DNA replicating enzymes.
Introduction
The genetic (hereditary) material for all living things is composed of DNA (deoxyribonucleic acid). The structure of DNA must enable this substance to store coded information that control the biological function of cells. The genetic material transmits this hereditary information in a stable form for the cell and organism through accurate replication of DNA. Although DNA is capable of change (we will discuss how this happens when we talk about DNA mutations), the replication process ensures high accuracy in copying the genetic information so that all progeny cells receive the same information. The DNA from all chromosomes in a human cell would be more than 6.5 feet long! Therefore, to fit inside the nucleus of a cell, DNA is packaged into chromosomes.
Read this article about Watson and Crick for more insight on the experimental facts discovered by chemists and biologist which contributed to the determination of the structure of DNA.
Chemical Structure of DNA Subunits
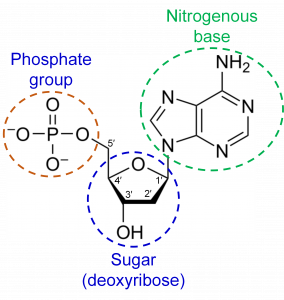
DNA is a polymer made of nucleotide subunits. A nucleotide consists of 3 chemical groups; a sugar, a phosphate and a nitrogenous base (Figure 1). In the case of DNA, the sugar is deoxyribose.
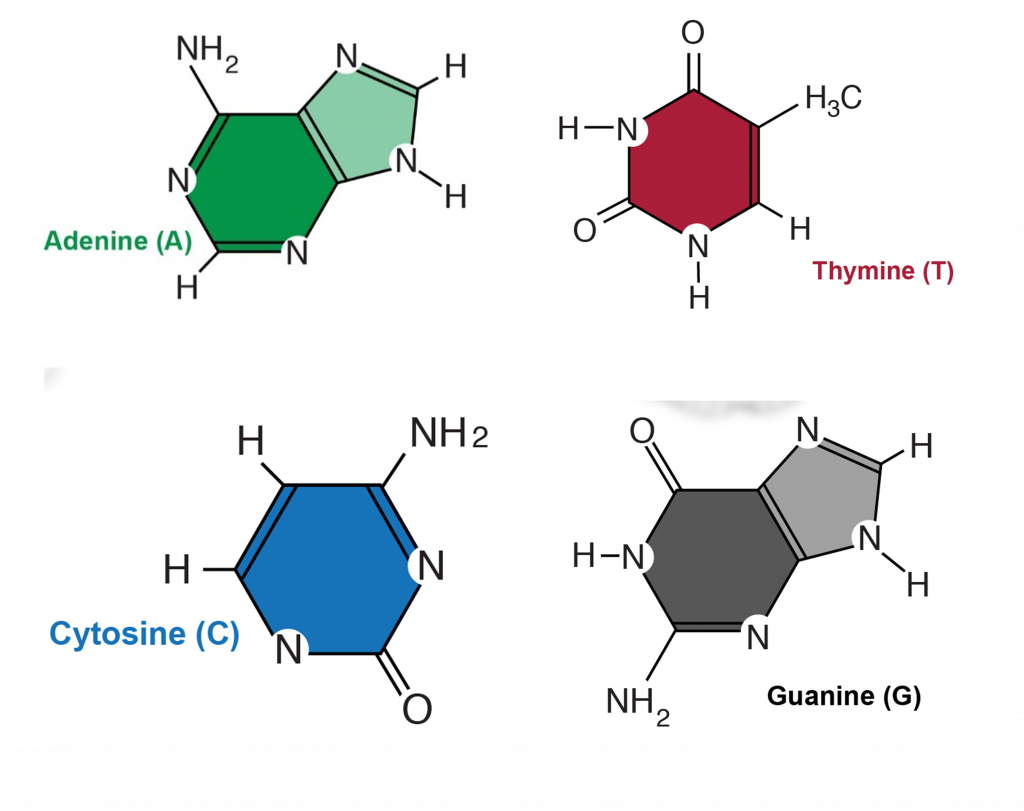
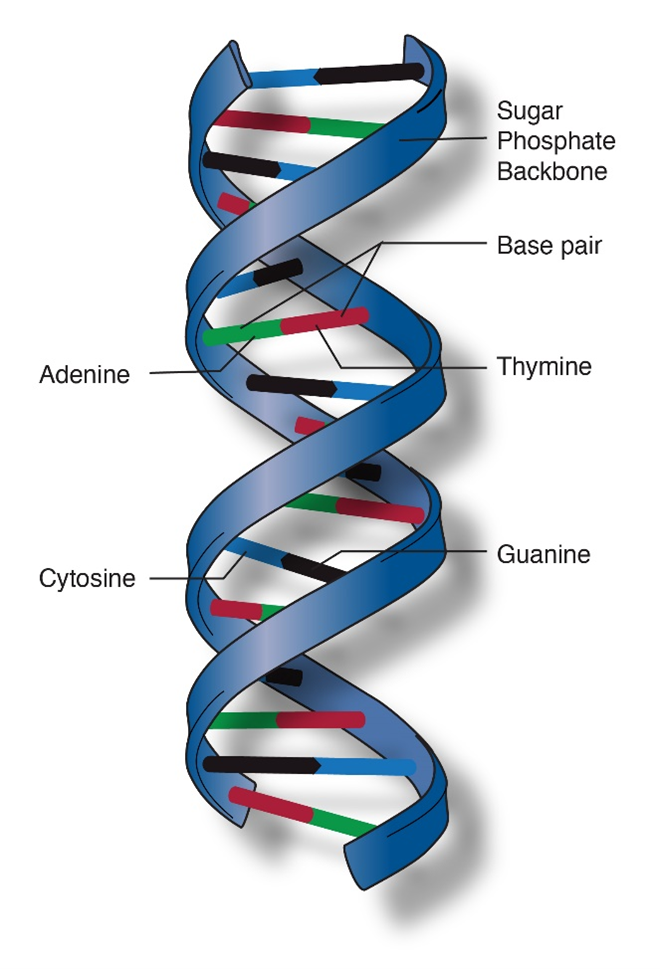
There are 4 different nucleotides in DNA that contain 4 different bases (Figure 2). These bases are named adenosine (A), cytosine (C), guanine (G) and thymine (T).
Complementary, antiparallel DNA strands
DNA codes information by ordering the sequence of the A,T,G and C nucleotides in a long polymer. The nucleotides are connected by phosphate bonds to form a strand. Two strands form a double stranded molecule (Figure 3). Note that in Figure 1, the carbon atoms on the deoxyribose sugar are numbered. The phosphate bond connects carbon #3 (the 3 prime or 3′ carbon) of one sugar to the 5′ carbon of the next. This gives a DNA strand directionality because the 5′ carbon faces one way and the 3′ carbon faces the other.
The bases on one strand form hydrogen bonds with the bases on the other. This is called “base pairing” and it is very specific, A only pairs with T and C only pairs with G. Thus, the sequences on the two strands of DNA are “complementary”; whenever there is an “A” on one strand, there is a “T” in the corresponding position of the complementary strand.
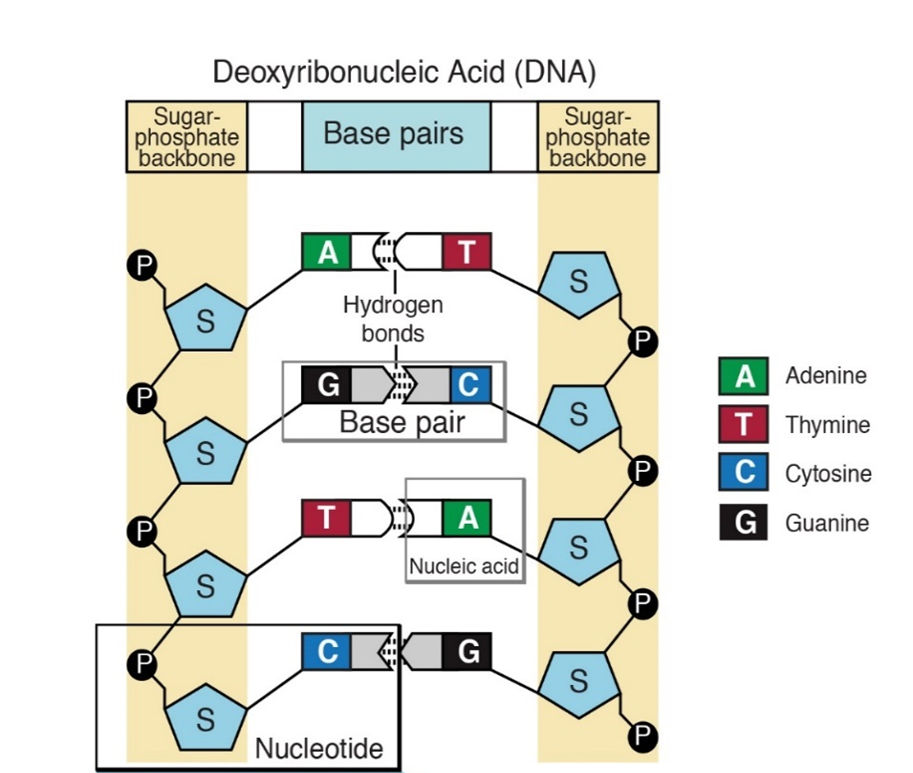
In addition to containing complementary nucleotide sequences, the strands are in opposite orientations. The 5′ end of one strand is oriented toward the 3′ end of the other. For this reason, the strands are referred to as “antiparallel”.
Double helix
The final feature of the molecular structure is that DNA assumes a helical conformation (Figure 4). This is the most stable configuration that accommodates all the molecular structures and chemical bonds that make up DNA.
Given the following sequence of a DNA strand, predict the sequence and orientation of the complementary strand.
5′-G-A-C-C-G-T-A-A-T-C-G-C-3′
Answer: 3′-C-T-G-G-C-A-T-T-A-G-C-G-5′
Packaging of DNA into chromatin and chromosomes
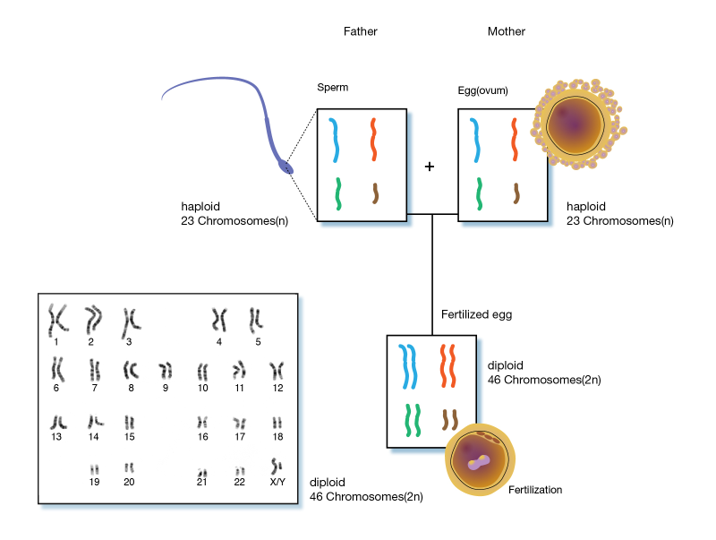
Chromosomes are long double stranded DNA molecules. Over 150,000,000 nucleotide pairs make up the human X chromosome. The complete replication and orderly transfer of something this large requires the chromosome to be packaged for stability and organization which creates genetic stability in the cells of a species. Every cell in a diploid organism contains two copies (2n) of every chromosome present in that organism (Figure 5). For example, humans have 46 chromosomes in their body, 23 were inherited from the father and 23 from the mother. Gametes, the reproductive cells of an organism, (egg or sperm), have only one set (1n) of chromosomes. When the two gametes unite, they form a living embryo with two sets of genetic information. Therefore, we actually have two copies of the genetic information for each trait. Sometimes one copy controls trait expression, and other times both copies influence a trait. As a result, the offspring will have characteristics of both the mother and the father.
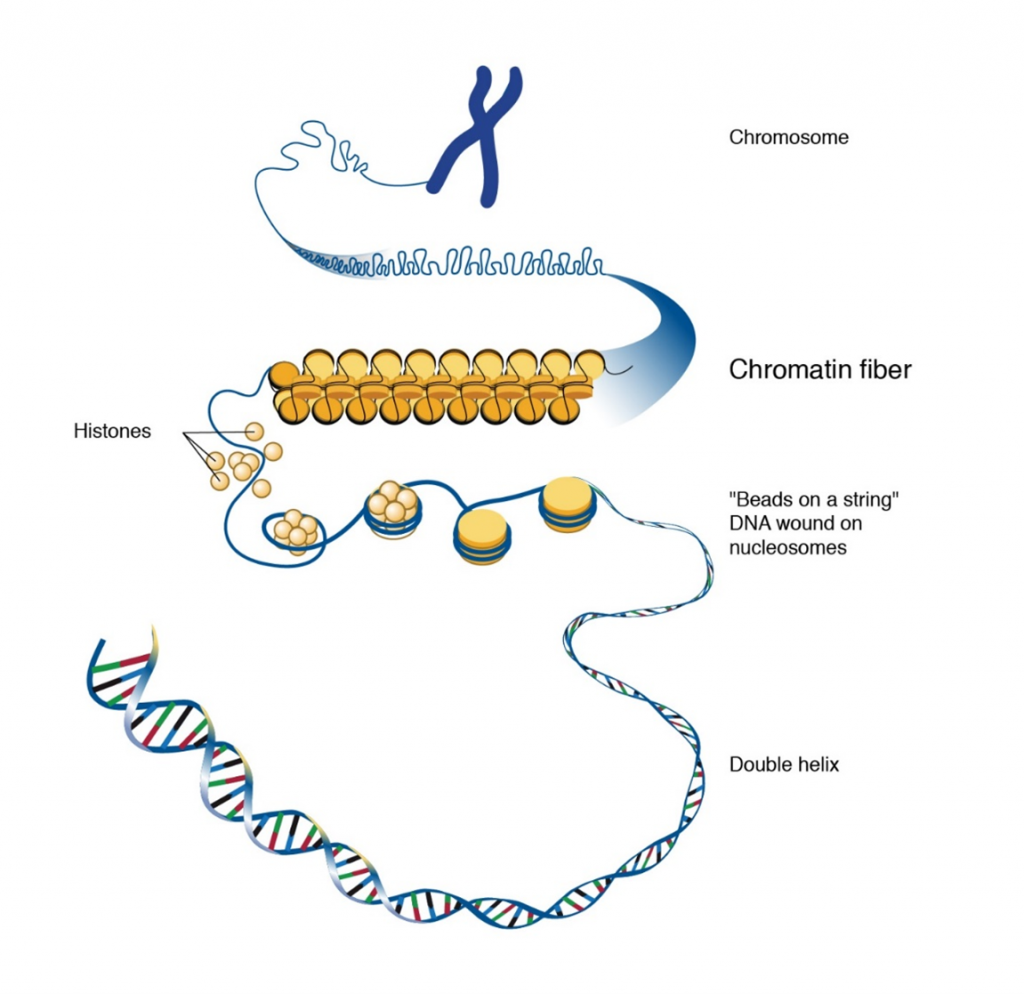
Inside cell nuclei, DNA is assembled, or packaged, into material called chromatin, which is composed of both DNA and proteins (Figure 6). Chromatin functions to protect and regulate DNA, as well as to efficiently store the very long DNA molecules that make up chromosomes within the limited space of the nucleus.
DNA from higher eukaryotic organisms is about 109 to 1010 bp. Human and maize chromosomal DNA is about 109 bp, which is equivalent to 1.8 m if all chromosomal DNA were stretched end to end in a linear manner. The diameter of the nucleus is only about 4-6 μm making it a challenge to fit chromosomal DNA inside the cell nucleus. This is accomplished by the ability of DNA to assume a very condensed structure to fit inside the nucleus. Therefore, the organization of eukaryotic DNA into chromatin is an important aspect of DNA packaging.
Nucleosomes and histones
The ordered coiling of chromosomal DNA around a histone protein core forms the chromatin. Chromatin is made up of nucleosomes (Figure 6) which represent the association of chromosomal DNA with histone proteins. A nucleosome is made up of about 145-147 base pairs of DNA coiled around each histone octamer, for about two complete turns. A histone octamer consists of two copies of each core histone. The total mass of the histones in the nucleus approaches that of the DNA – 2 molecules of each core histone to approximately 200 bp of DNA.
Higher order structures
If the higher order chromatin structure is disrupted, electron microscopy reveals the appearance of “beads on a string” with a diameter of about 10 nm. The “beads” represent the DNA wrapped around histones. Nucleosome formation results in a DNA fiber that is about 10 nm and a packaging ratio of about 7. The higher order chromatin structure results when the 10 nm fiber is coiled into a solenoid. The result of nucleosome coiling is a chromatin fiber of 30 nm that is observed by electron microscopy.
Centromeres
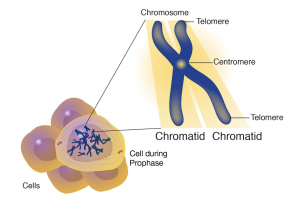
The centromere is a chromosomal region that controls chromosome segregation at mitosis and meiosis (Figure 7). Centromeres connect to microtubules of the spindle apparatus, which directs their movement to opposite poles (daughter cell nuclei) during cell division. Scientists were successful in isolating these centromeric sequences from yeast and engineered brewer’s yeast (Saccharomyces cerevisiae) plasmids that were able to replicate like the chromosomes. Through these efforts scientists were able to point centromeric function to a DNA stretch of about 120 bp that was resistant to DNAse and bound to a single microtubule.
Telomeres
The telomere lies at the end of the chromosome and confers stability by “sealing” the end of a chromosome (Figure 7). Telomeres consist of long series of short repeated DNA sequence that occurs in tandem arrays and are added to the end of the chromosome during DNA replication by an enzyme called telomerase. In most plant species the sequence TTTAGGG constitutes a conserved telomere motif.
The knowledge about the structure and function of centromeres and telomeres has had a profound impact in molecular genetics. For example, yeast artificial chromosomes (YACs) and bacterial artificial chromosomes (BACs) have proven useful in the physical mapping of plant genomes requiring the cloning and multiplying large (>100 kb) DNA fragments in yeast or bacterial cells.
DNA Replication
In 1953, J.D. Watson and F.H.C. Crick published a note in NATURE; one of the world’s most widely read research journals. The note cited just six references.
Watson and Crick’s writing had limited chemistry details given the title of their note, “A Structure for Deoxyribonucleic Acid”. A later article would share more of the chemistry behind their proposed double helix structure for Deoxyribonucleic Acid (DNA). This note in Nature was written for a broader audience, including biologists who recognized that the structure of DNA was a prerequisite for understanding the function of the genetic material. They included a diagram to allow readers to visualize this unique double helix structure. To emphasize the structure determines function importance, Watson and Crick included this statement in their note…
“It has not escaped our notice that the specific pairing we have postulate immediately suggests a possible copying mechanism for the genetic material.”
Accurate replication prior to cell division is one of the three key functions of the genetic material. While later experiments would reveal the details of how cells replicate DNA, Watson and Crick were obviously impressed that the proposed double helix structure would fit with the concept that living cells are needed to produce more living cells and the transmission of a complete set of biological instructions. The genetic material must have a structure that lends itself to being copied. Using old to help build new leads to a proposed semiconservative model for replicating the double stranded DNA molecule (Figure 8).
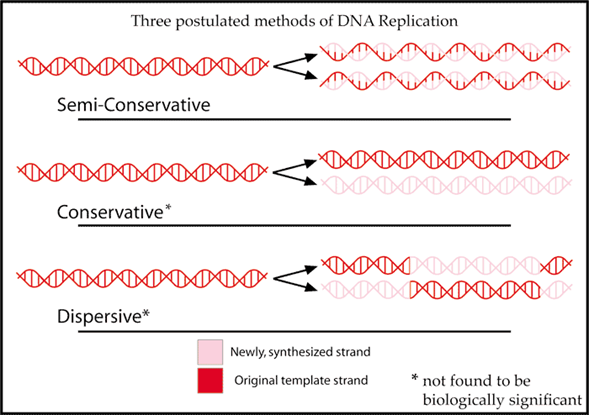
Prior to both mitosis and meiosis cell division in multicellular organisms, and cell division in prokaryotes, the DNA inside the cell must be replicated. (see lesson Mitosis and Meiosis). This replication process generates the genetic information needed for either two cells, genetically identical with the original cell or in sexually reproducing organisms, four gamete cells with half of the original cell’s genetic information.
When and Where:
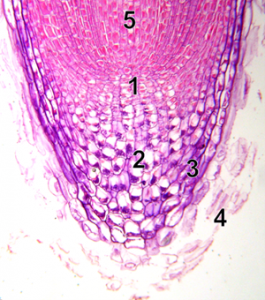
DNA replication will happen before cells divide. In hours old embryos that are developing into multicellular organisms, all the cells are replicating their DNA and dividing into new cells. Hours or days later, multicellular organisms have developed specific meristem regions where new cell production occurs. A visual example of meristem cells where DNA replication is needed is the tip of a plant root (Figure 9). New cells made at the root tip allow the root to grow. The root must also replace the cells that are damaged as the growing root moves through soil. In Figure 8, the meristem region (1) contains meristem cells that are replicating their chromosomes, then dividing into two identical cells by mitosis. Cells on the tip part of the meristem specialize in root cap cells (2,3) with the job of protecting the meristem. The fate of these cells is to die (4), and they need to be replaced by new cells made in the meristem. Cells made on the other half of the meristem (5) can elongate and eventually specialize. These root cells will then function for the plant, all season for annual plants, years for perennial plants. Replication of DNA will only happen in the meristem cells.
Speed of chromosome replication
Fifty-two years after Watson and Crick’s Nature note, a group of about 50 molecular geneticists published the complete DNA sequence of the human X chromosome (Figure 5). This discovery revealed that X chromosome is one double-stranded DNA molecule that is about 155 million nucleotide pairs long. Every time a human cell divides, one or two of these 155,000,000 nucleotides must be replicated. This must be done with speed and accuracy.
The main DNA replication enzyme (DNA polII, see below) works fast. Estimates of its replicating speed are around 750 nucleotides per second.
Let us do the math.
155,000,000 nucleotides / chromosome X 1 second / 750 nucleotides
= 206,666 seconds per X chromosome
Let us convert these seconds to hours.
1 hour / 360 seconds X 206,666 seconds/chromosome = 547 hours per chromosome
That means one replication enzyme would take 547 hours or about 24 days to replicate an X chromosome. What are the biological ramifications of this 24 days? If you cut yourself, it would take 24 days to replicate the chromosomes before the skin cells could divide. The formation of new cells needed for healing cuts in our skin would keep us in a wounded state.
To speed up the chromosome replication process, two tactics are used by living cells. Both tactics are shown in Figure 10. First, replication will not start on just one end of the chromosome. Instead, there are hundreds of origins of replication (ori) along the chromosome. In addition, the replication process moves bidirectionally from the ori. This bidirectional replication creates forks of replication that move toward each other as the chromosome is replicated.
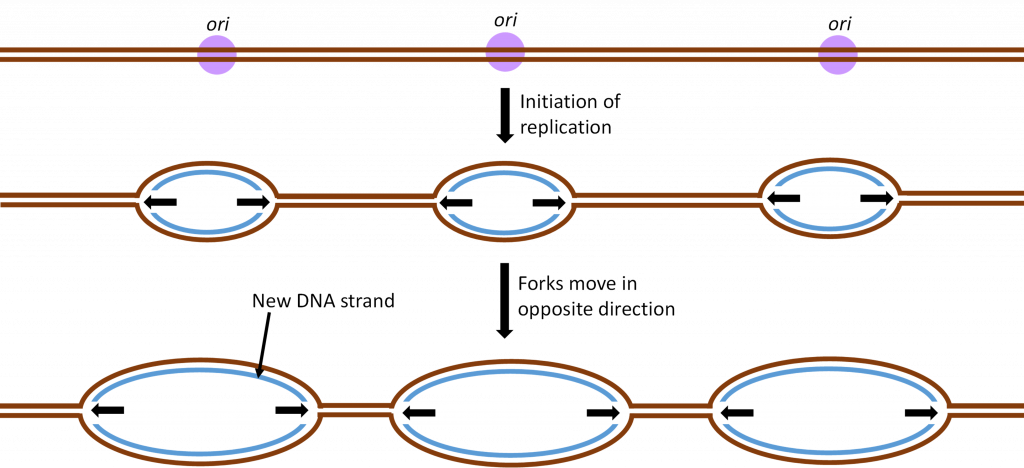
As a result of these two tactics, a large chromosome can be replicated in hours rather than weeks and cell division can happen quickly when it is needed for growth or cell replacement.
The Chromosome Replicating Team
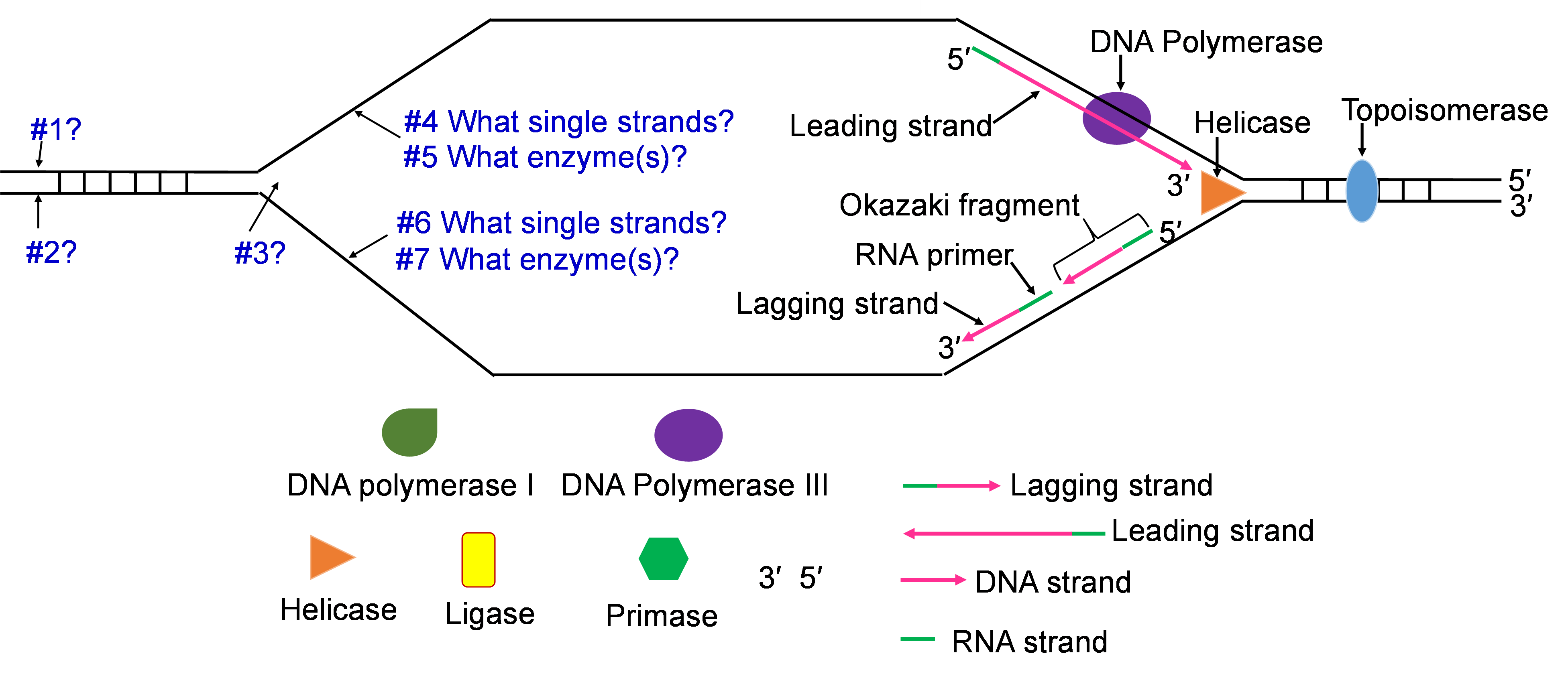
Like all processes in living cells, a collection of specific proteins works together to perform functions which control and complete chromosome replication. All the proteins involved in DNA replication make and break bonds, so they are enzymes. The function of the main DNA replicating enzymes is described below.
Five of the main DNA replicating enzymes
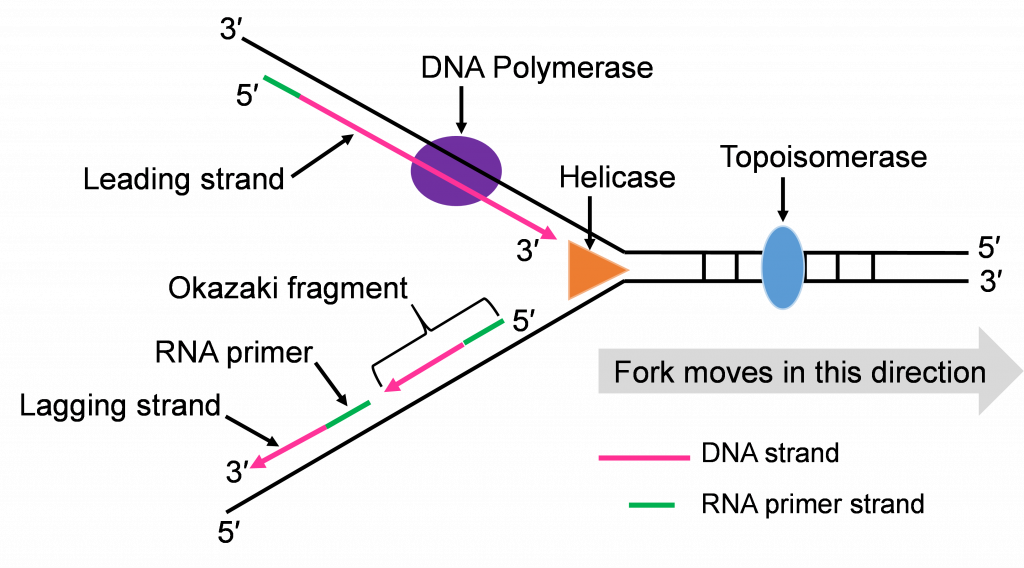
- Helicase: This is one of the enzymes that unwinds the double stranded DNA molecule. The unwound single strands no longer hydrogen bond to their complementary strand but the sugar-phosphate bonds remain intact.
- Topoisomerase: The unwinding work of Helicase creates tension in the double stranded DNA ahead of the replication fork. Topoisomerase relieves this tension by catalyzing a series of sugar phosphate bond breaking and making steps. Without this tension relief, the Double stranded DNA could break and the intact chromosome cannot finish replicating.
- DNA Polymerase III (DNA pol III): This is the main DNA synthesizing enzyme. The enzyme reads the single strand as a template and places in the complementary deoxyribonucleotide. DNA pol III reads the template in the 3′ to 5′ direction and builds the new strand in the 5′ to 3′ direction, adding the next nucleotide to the 3′ end by catalyzing sugar-phosphate bonding. DNA pol III illustrates the specificity of enzymes several ways; it reads and proofreads the placement of new nucleotides to insure accurate replication. It can only read and build in one direction. DNA pol III can only add nucleotides to a free 3′ end. This last specification means that DNA pol III cannot start the replication process on the single stranded template. Another enzyme needs to be part of the in vivo replication team to prime the work of DNA pol III.
- Primase: Biochemists named this enzyme to describe its role in starting or priming the replication process. DNA (or RNA) primase is a special RNA polymerase. The enzyme reads the single stranded DNA template 3′ to 5′ and adds ribonucleic acid (RNA) nucleotides in the 5′ to 3′ direction. Once a few hundred RNA nucleotides are added, primase falls off the template strand and leaves the 3′ end that DNA Pol III needs to continue the process.
- DNA Polymerase I (DNA pol I): The DNA polymerase specializes in the removal of the RNA primers and replacing them with DNA nucleotides. The enzyme works with the same 3′ to 5′ reading and 5′ to 3′ building.
- DNA Ligase: After the five enzymes described above have completed their work, some sugar phosphate bonds need to be made to complete the double stranded molecule. DNA ligase has this backbone bond sealing assignment.
Now that we have introduced the replication enzymes, we can describe the step-by-step action of this team. We will focus on the action that happens at one origin of replication.
Step 1: Initiating replication at the Ori
Helicase enzymes bind to the ori sequences and start to unwind the double stranded DNA. This establishes two replication forks (one shown in Figure 10a) and a helicase enzyme will be working to unwind the DNA at each fork as the forks move away from each other. The unwinding exposes single strands of DNA that are the template for building a new strand. Topoisomerase can be seen working to relieve tension ahead of the replication fork in Figure 10a.
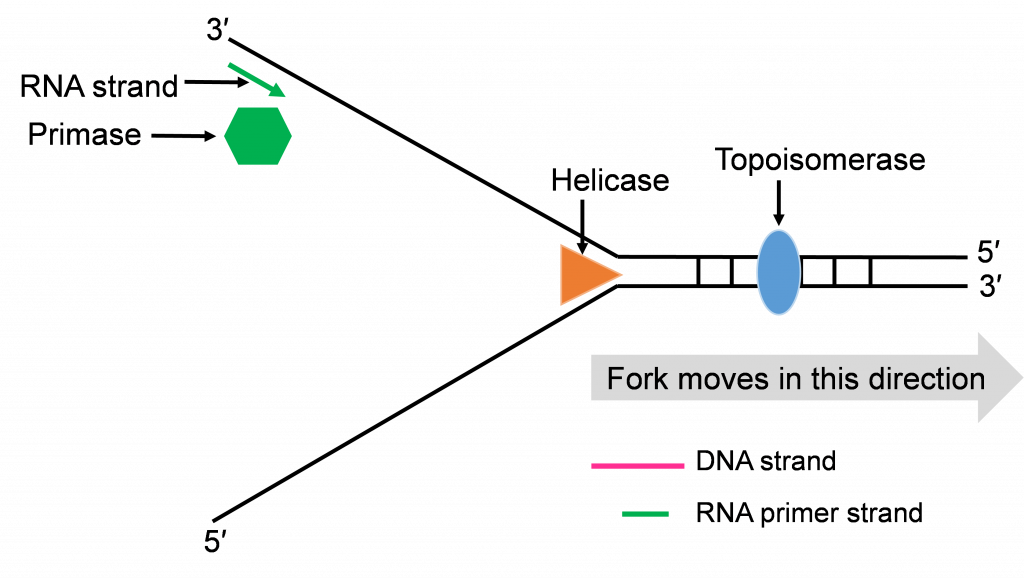
Step 2: Priming DNA with RNA
The process of priming with primase and then replicating with DNA pol III happens at both replication forks (Figure 10a and b). DNA pol III enzymes can replicate new strands as fast as the DNA is unwound at each replication fork. The unwinding creates two template strands at each fork so primase enzyme must work to prime both old strands.
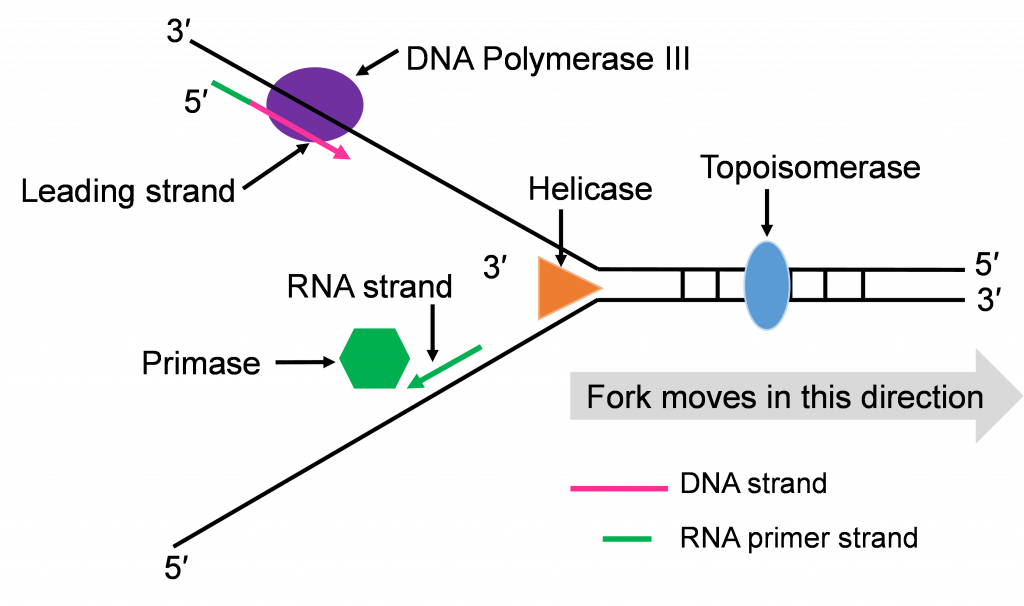
Step 3: Synthesis of leading and lagging strands
The double strands of DNA are anti-parallel and ‘run’ in opposite directions and DNA pol III can only work in one direction (Figure 10c). Thus, there is continuous and discontinuous replication happening at each replication fork. The strand that can be replicated as helicase unwinds is the leading strand. The DNA pol III must replicate the other strand in the opposite direction so this strand will lag behind. (Figure 10c).
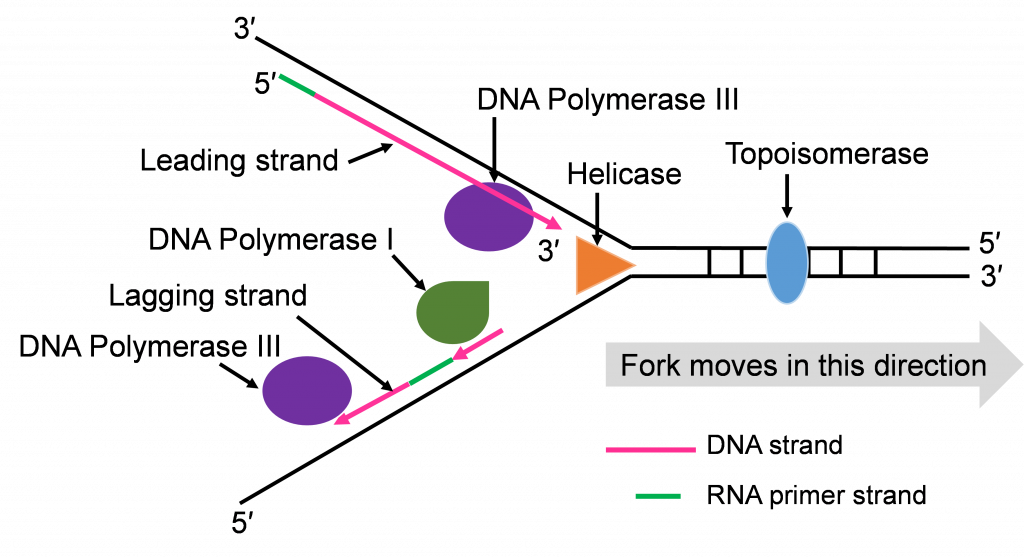
There is more priming needed to replicate the lagging strand and the priming leaves RNA-DNA hybrid stretches in the new double stranded molecule. This means there is work for the enzyme DNA pol I. This enzyme will be part of the removal of the RNA nucleotide primers, and replacement with DNA nucleotides (Figure 10c and d).
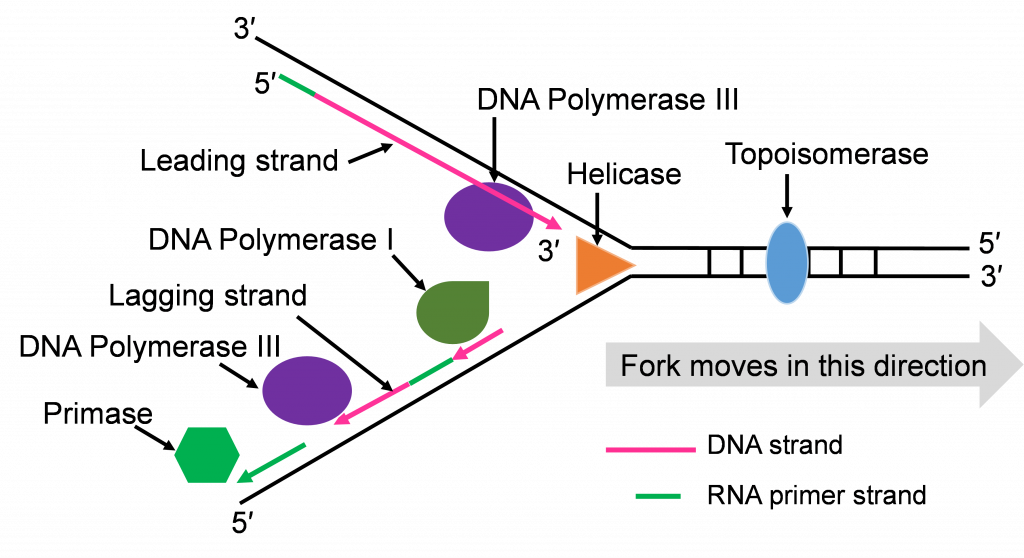
Both replication forks have a leading and a lagging strand of new DNA. The DNA nucleotide stretches between the red RNA primers would be the Okazaki fragments.
One of the first scientists to provide evidence to support this model of DNA replication was the team of Reiji and Tsuneko Okazaki in the 1960s. They used radioactive nucleotides to track the newly replicating fragments and found that many of the fragments were short. This fits with the discontinuous model.
As the four enzymes just described attend to their specific roles in replication, two double stranded DNA molecules with an old strand and a new strand are built. If the DNA pol III makes a mistake and adds the wrong, non-complementing nucleotide, the enzyme can proof read its work and replace these replication mistakes. When replication is perfect, the two double stranded DNA molecules will have identical sequences.
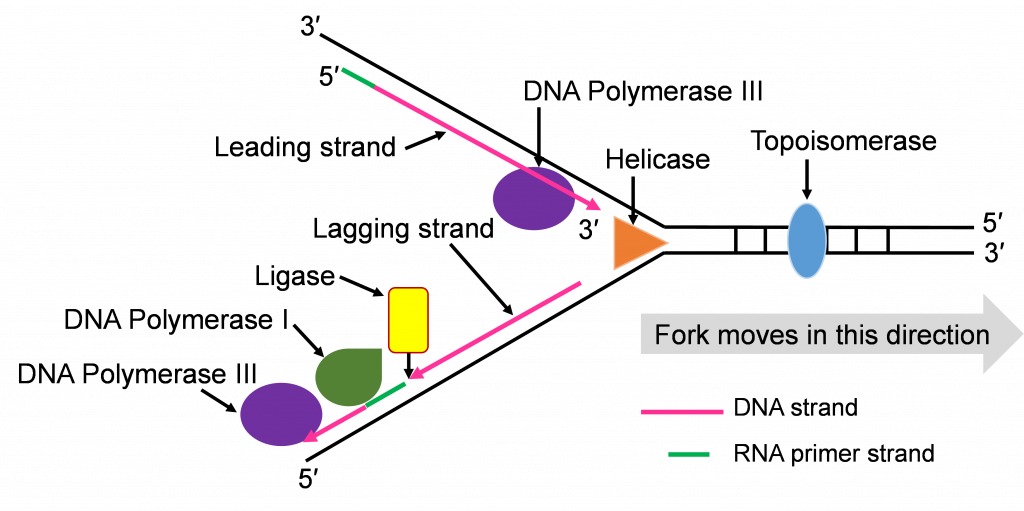
Step 4: Ligation of Okazaki fragments
The fifth enzyme that has a final role in completing replication is DNA ligase (not shown in Figure 10d and e). This enzyme will seal the sugar phosphate bond between the last replacement nucleotide added by DNA pol I and the first nucleotide that had been added after priming by DNA pol III. With this bond sealing work, the replication process is complete.
The two double stranded DNA molecules that get made will have identical sequences unless rare mistakes happen as the enzymes conduct their work.
DNA is composed of nucleotides. Nucleotides are connected together by phosphate bonds to form a strand. The bases on one strand form hydrogen bonds with the bases on the other to form a double stranded molecule. The final feature of the molecular structure is that DNA assumes a helical conformation. To fit inside the nucleus, DNA assumes a very condensed structure. Therefore, chromosomal DNA is coiled around a histone protein core to form chromatin. The tight packaging of DNA in chromatin must be modified to allow DNA replication and transcription. In the process of replication, the two DNA strands separate and act as templates for the synthesis of complementary daughter strands.
Place the single strands, replication enzymes, and 3′ / 5′ labels in the appropriate places (#1-#7)