1.6: Gene Expression- Translation
- Page ID
- 73671
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Determine the amino acid sequence of a protein given the nucleotide sequence of a gene.
- Recognize the biological connection among mRNA, tRNA, amino acids, and proteins.
- Understand the concept of protein structure and its relation to protein function.
Introduction
The importance of gene expression is evident when you observe the changes plants go through during their lifecycle or during a season. Trees and bushes, for example have dormant buds through the winter. Environmental signals affiliated with the coming of spring induce genes in the buds to turn on and drive the dramatic changes of leaf development and flowering. The genes were always present in those bud cells but were controlled to turn on at the proper time. Understanding gene expression thus requires an examination of two processes, the activation of gene expression to make a “message” and the reading of this message to build a specific protein.
Amino acids are used to make proteins
Amino acids are the building blocks of proteins. Except for the amino acid proline, they all consist of a central carbon atom covalently bonded to an amino group, a hydrogen atom, a carboxyl group, and a variable R group (Figure 1). In the physiological range, both the carboxylic acid and amino groups are completely ionized allowing amino acids to act as either an acid or a base. Thus, amino acids do not assume a neutral form in an aqueous environment.
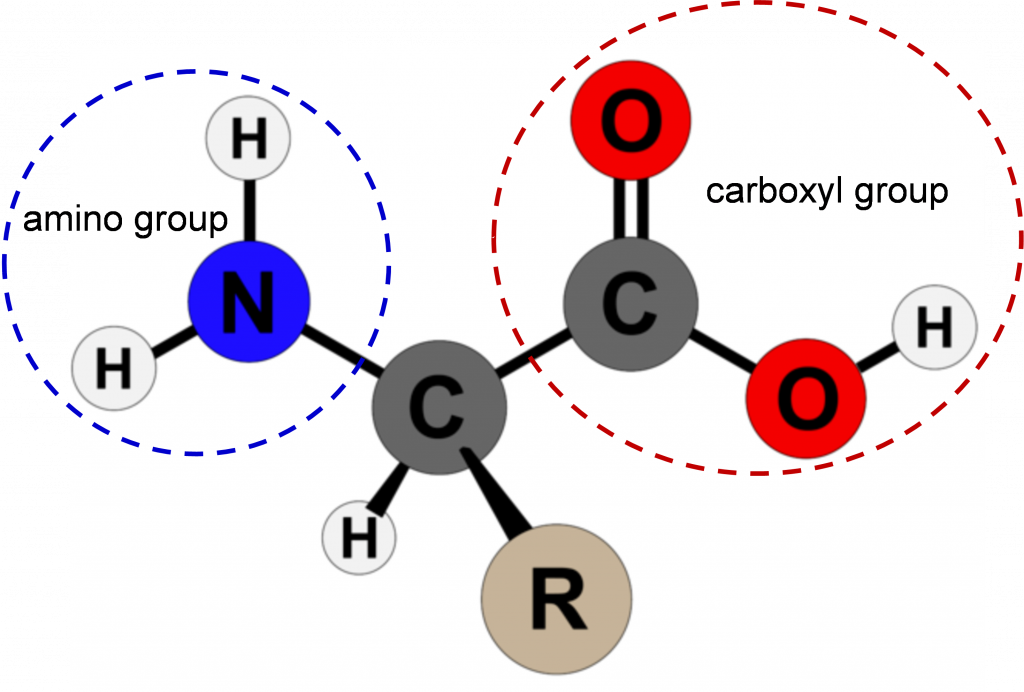
The chemical and physical properties of the side chains provide the functionally important properties to amino acids. Some of the important properties of side chains include acidic, basic, or neutral, hydrophobic vs. hydrophilic, as well as the size of side chains. They are categorized according to their side groups as nonpolar, polar, positively, or negatively charged or aromatic. All these properties and others affect how each amino acid contributes to the structure and function of a mature protein.
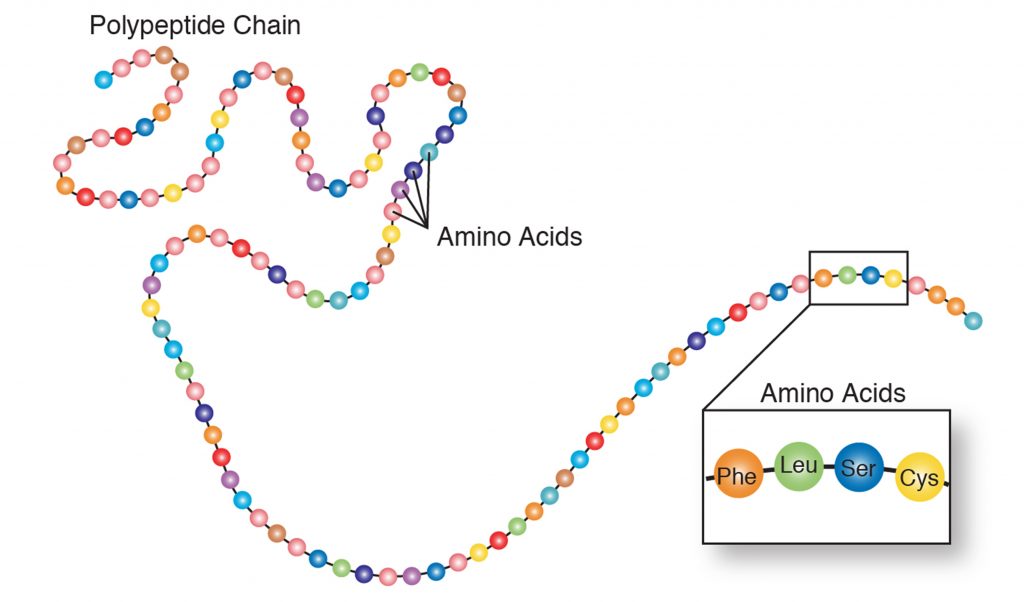
There are 20 different amino acids making up the subunits of proteins (Table 1). All proteins are made from differing combinations of the amino acids (Figure 2). Therefore, if you are going to make proteins you need to either make amino acids first (Figure 3) or consume amino acids in your diet. Animals consume some of their amino acids (the essential amino acids), but plants and bacteria are able make all their own amino acids.
| Full Name | Abbreviation (3 Letter) | Abbreviation (1 Letter) |
|---|---|---|
| Alanine | Ala | A |
| Arginine | Arg | R |
| Asparagine | Asn | N |
| Aspartate | Asp | D |
| Cysteine | Cys | C |
| Glutamate | Glu | E |
| Glutamine | Gln | Q |
| Glycine | Gly | G |
| Histidine | His | H |
| Isoleucine | Ile | I |
| Leucine | Leu | L |
| Lysine | Lys | K |
| Methionine | Met | M |
| Phenylalanine | Phe | F |
| Proline | Pro | P |
| Serine | Ser | S |
| Threonine | Thr | T |
| Tryptophan | Trp | W |
| Tyrosine | Tyr | Y |
| Valine | Val | V |
Codons and the Genetic Code
The search for the genetic code (Table 2) revealed that genetic information is stored in nucleotide triplets referred to as codons. The genetic code is degenerate because many amino acids are specified by more than one codon. Sixty one of the 64 possible combinations of the three bases in a codon are used to code for specific amino acids. Three stop codons, UAA, UAG, and UGA do not code for any amino acids but specify the termination of peptide chain synthesis during translation. The AUG start codon is used to initiate polypeptide synthesis and codes for the amino acid methionine.
|
First Position |
Second Position |
Third Position |
|||
|
U |
C |
A |
G |
||
|
U |
Phe (F) |
Ser (S) |
Tyr (Y) |
Cys (C) |
U |
|
C |
|||||
|
Leu (L) |
Stop |
Stop |
A |
||
|
Stop |
Trp (W) |
G |
|||
|
C |
Leu (L) |
Pro (P) |
His (H) |
Arg (R) |
U |
|
C |
|||||
|
Gln (Q) |
A |
||||
|
G |
|||||
|
A |
Ile (I) |
Thr (T) |
Asn (N) |
Ser (S) |
U |
|
C |
|||||
|
Lys (K) |
Arg (R) |
A |
|||
|
Met (M) |
G |
||||
|
G |
Val (V) |
Ala (A) |
Asp (D) |
Gly (G) |
U |
|
C |
|||||
|
Glu (E) |
A |
||||
|
G |
|||||
Why a Triplet Code?
Prior to understanding the details of transcription and translation, geneticists predicted that DNA could encode amino acids only if a code of at least three nucleotides was used. The logic is that the nucleotide code must be able to specify the placement of 20 amino acids. Since there are only four nucleotides, a code of single nucleotides would only represent four amino acids, such that A, C, G and U could be translated to encode amino acids. A doublet code could code for 16 amino acids (4 x 4). A triplet code could make a genetic code for 64 different combinations (4 X 4 X 4) genetic code and provide plenty of information in the DNA molecule to specify the placement of all 20 amino acids. When experiments were performed to crack the genetic code, it was found to be a code that was triplet. These three letter codes of nucleotides (AUG, AAA, etc.) are called codons.
The genetic code only needed to be cracked once because it is universal (with some rare exceptions). That means all organisms use the same codons to specify the placement of each of the 20 amino acids in protein formation. A codon table can therefore be constructed and any coding region of nucleotides read to determine the amino acid sequence of the protein encoded. A look at the genetic code in the codon table below reveals that the code is redundant meaning many of the amino acids can be coded by four or six possible codons. The amino acid sequence of proteins from all types of organisms is usually determined by sequencing the gene that encodes the protein and then reading the genetic code from the DNA sequence
Ribosomes, tRNA and Anti-codons
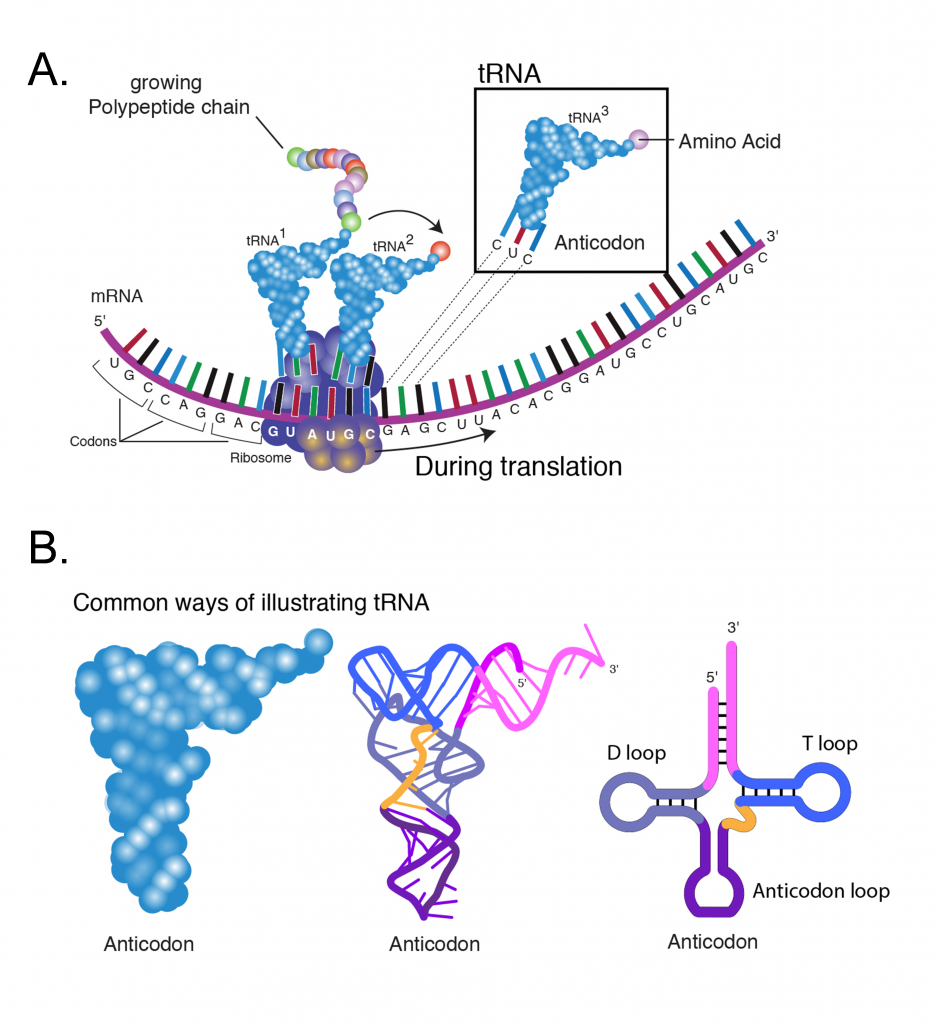
The ribosomes are a large molecular complexes containing an assembly of several ribosomal RNAs (rRNA) and many proteins. Ribosomes are the machinery of protein synthesis, facilitating the ordered addition of amino acids to a nascent polypeptide chain under the guidance of an mRNA template (Figure 5A). Prior to the initiation of protein synthesis the ribosome occurs in two separate subunits of size 60s and 40s (“s” stands for Svedberg units and is a measure of a particles sedimentation rate during centrifugation).
The meaning of a codon for a specific amino acid is determined by the tRNA (transfer RNA). Each tRNA (Figure 5B) contains a triplet of nucleotides referred to as an anticodon, which is complementary to a specific codon. For each of the 20 amino acids, a specific enzyme (aminoacyl-tRNA synthetase) catalyzes its linkage to the 3′ end of its specific tRNA adapter. The function of aminoacyl-tRNA synthetase is critical as it provides a check for accuracy in protein synthesis by adding a specific amino acid to a specific tRNA molecule. In this way, one particular amino acid is targeted to each codon triplet of mRNAs.
Peptide chain formation
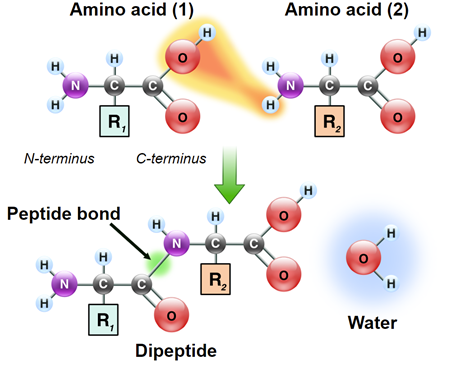
Protein synthesis is initiated by the 40s subunit through attachment it’s to the mRNA, and recognition of the AUG codon. This is followed by the attachment of the 60s subunit to the 40s-mRNA complex to provide the structure necessary to align each successive aminoacyl-charged tRNA for transfer of its amino acid to the growing polypeptide. Successive amino acids are attached to the growing polypeptide by the formation of peptide bonds that form between the carboxyl group of one amino acid and the amino group of the next.
Like nucleic acids, polypeptides also have molecular directionality. One end of a polypeptide chain will have a free amino group and the opposite end will have a free carboxy group. As such, these ends are referred to as the amino-terminus or carboxy-terminus, respectively. During translation, mRNA templates are read from the 5’ end toward the 3’ end. Proteins are synthesized beginning at the amino terminal end going toward the carboxy terminus.
Protein structure and function
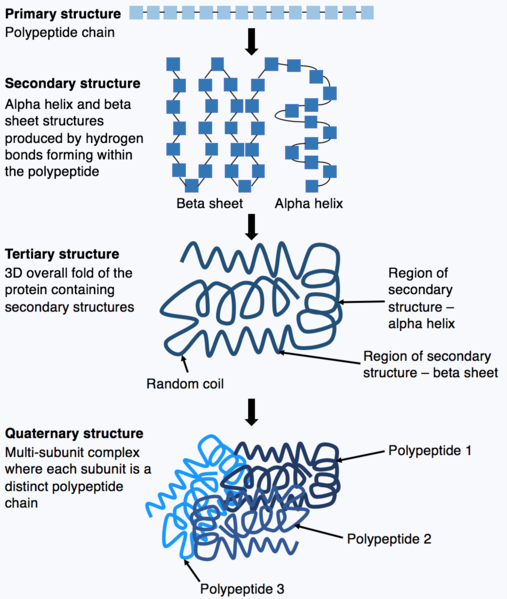
Proteins assume complex 3-dimensional structures that are essential for their various functions as enzymes, regulatory factors, or structural proteins. Overall structure is determined by complex physico-chemical interactions among amino acid side groups.
Several levels of structure are considered (Figure 6). The primary structure of a protein is the amino acid sequence of its polypeptide chains. The secondary structure is derived through the interactions between neighboring amino acids to form local structural elements such as beta sheets or alpha helices. The tertiary structure refers to the three-dimensional structure of an entire polypeptide and is determined by the diverse properties of amino acid side groups. For example, in an aqueous environment, hydrophobic amino acids will often interact at the core of a protein structure while hydrophilic groups may be exposed on the protein surface.
Many proteins are multimeric, composed of several polypeptide chains called “subunits”, which associate non-covalently or in some cases via disulphide bonds. Such higher order spatial associations among polypeptides are also the result of interactions among amino acids and result in a quaternary structure. Some multimeric proteins consist of multiple copies of the same polypeptide.
The genetic information of DNA is transferred to through transcription to an intermediate molecule called RNA. The signals for starting and stopping transcription are located within the DNA sequence, and referred to as promoter and terminator sequences. The coding region of a gene is composed of a sequence of nucleotides that are transcribed into RNA. These sequences include exons and introns. Exons are the sequences that code for proteins. The coding region of a gene contains exons and introns. Also, pre-mRNA contains both introns and exons. The introns in pre-mRNA are removed through a process called intron splicing. The mRNA is processed by 5’ capping and addition of a poly(A) tail. Mature RNA is then translated to amino acids used to build proteins.
- Table Source: Donald Lee, University of Nebraska-Lincoln ↵


