18.3: Nitrogen Excretion and the Urea Cycle
- Page ID
- 15033
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)-
Explain the Overall Role of the Urea Cycle:
Understand how the urea cycle detoxifies excess ammonia generated from amino acid catabolism and why urea is a preferred end product for nitrogen excretion. -
Describe the Key Enzymes and Their Functions:
Identify and explain the roles of the main enzymes (CPS1, OTC, ASS1, ASL, ARG1) in the urea cycle, including their specific reaction steps and contributions to overall cycle function. -
Analyze Enzyme Mechanisms and Structural Features:
Evaluate the reaction mechanisms of enzymes like carbamoyl phosphate synthetase I and arginase, including details such as ATP consumption, active site architecture, substrate tunnels, and the role of cofactors (e.g., N-acetyl-glutamate). -
Discuss Regulation and Allosteric Control:
Understand how key regulators, such as N-acetyl-glutamate, control the rate-limiting step of the urea cycle, and explore how enzyme activity is modulated in response to metabolic demands and dietary protein levels. -
Examine Experimental Evidence for a Cyclic Pathway:
Assess how experiments—like the addition of ornithine—demonstrate the cyclic nature of the urea cycle and the concept of catalytic regeneration within the cycle. -
Connect the Urea Cycle to Broader Metabolism:
Explore how the urea cycle is linked to the tricarboxylic acid (TCA) cycle, particularly through intermediates such as fumarate and aspartate, and understand the role of metabolic shuttles (e.g., malate–aspartate shuttle) in integrating these pathways. -
Evaluate Energetic and Metabolic Implications:
Quantify the energy cost of urea synthesis (in terms of ATP expenditure) and discuss how the cycle’s integration with the TCA cycle helps compensate for this energetic demand. -
Apply Structural Biology to Enzyme Function:
Utilize interactive molecular models to gain insights into enzyme structure, such as the tunnel formation in CPS I or the multimeric organization of argininosuccinate lyase, and connect these structures to their catalytic functions. -
Interpret Clinical Relevance and Disorders:
Link the biochemical details of the urea cycle to clinical scenarios, such as hyperammonemia, and discuss how defects in specific enzymes can lead to metabolic disorders, highlighting the importance of enzyme localization and gene regulation. -
Compare Cyclic versus Linear Metabolic Pathways:
Contrast the urea cycle with linear metabolic processes by examining the efficiency, regulation, and “recycling” nature of cyclic pathways, drawing parallels with broader concepts like the circular economy.
These goals aim to integrate mechanistic understanding, regulatory insights, structural biology, and clinical applications, providing a comprehensive framework for mastering the urea cycle and its connections to overall metabolism.
The Urea Cycle
Now, we can see what happens to excess NH3/NH4+ that accumulates in the liver mitochondria. The ammonia formed in the liver through oxidative deamination by glutamate dehydrogenase ends up in urea. Alternatively, it can be added to glutamic acid to form glutamine, which can be transported to the kidney. There, after sequential actions of glutaminase (a deamidation reaction) and glutamate dehydrogenase, it forms ammonia for direct excretion in the urine. If your diet is high in proteins and hence amino acids, since they can't be stored as proteins per se, they are metabolized for energy, and the metabolic products can be converted to fat or carbohydrates. That leaves excess nitrogen, which is mainly excreted as urea.
Urea is a highly water-soluble, non-toxic molecule that biochemists use in the lab to chemically denature proteins (at concentrations of 3-6 M). Clinical blood urea concentrations are expressed as Blood Urea Nitrogen (BUN), which ranges from 7 to 20 mg/dL N, equivalent to 2.5 to 7.1 mM urea. Serum levels depend predominantly on the balance between urea synthesis in the liver and its elimination by the kidney. Normal cellular urea concentrations should be similar.
Urea is produced through a cyclic pathway known as the urea cycle. You are familiar with perhaps the most favorite biochemical cycle, the tricarboxylic (citric or Krebs) cycle. Krebs also discovered the urea cycle before conceiving the citric acid cycle. He concluded that the urea formation pathway was a cycle using the same logic he deployed for the citric acid cycle. After your first view of the full urea cycle, we'll explain how, as shown in Figure \(\PageIndex{1}\).
Figure \(\PageIndex{1}\): The Urea Cycle
The enzyme names are:
- CPS1: carbamoyl phosphate synthase I
- OTC: ornithine transcarbamoylase
- ASS1: arginosuccinate synthase 1
- ASL: arginosuccinase lysase (aka arginosuccinase)
- ARG1: arginase
It is color-coded to designate the sources of the C and N atoms. One N comes from ammonia (or indirectly from the amido N of glutamine), while the C atom comes from carbonate. The other comes from the amine of aspartate. Note that three ATPs are used to power the cycle. Also note that the guanidino group of arginine (NH(NH2+)NH2) is the immediate donor of the 2 N atoms in the step that produces urea. Knowing that one amino acid was the immediate donor of the N atoms in urea, you would easily predict that.
Let's start with the first step, the cleavage of arginine to produce ornithine and urea. What happens to the ornithine derived as a "waste" product of urea formation? Experimentally, one might add more ornithine to a system running the urea formation pathway and ask what happens to the rate of urea synthesis. If it were a one-step process, you might expect, based on mass action, that adding ornithine, a product, would slow that reaction rate by driving it backward. Krebs found the opposite: adding more ornithine increased the rate of urea formation, but in a way that did not alter the ornithine concentration. Hence, ornithine was not consumed per se in a subsequent step. Ornithine appeared to act as a traditional catalyst, speeding up the reaction without being consumed. To do so, it must generate more arginine. The way to account for this was a cyclic pathway, not a linear or branched one. In a cycle, none of the participants is consumed in a net fashion.
Now, let's examine some of the steps, starting with those powered by ATP, which also incorporate nitrogen into the urea precursors.
Carbamoyl phosphate synthase I (CPS I)
This enzyme catalyzes the first "committed" step of the pathway and makes carbamoyl phosphate, the reactant that enters the cycle. It requires a specific cofactor, N-acetylglutamate (NAG), produced by the acetylation of glutamate by acetyl-CoA through the action of NAG synthase, which is activated by arginine. The reaction is not part of the cycle but provides input.
A cytosolic version of this enzyme, CPS II, synthesizes arginine and pyrimidine nucleotides using glutamine as a donor of NH4+, which reacts with carboxyphosphate to produce carbamoyl phosphate. NAG does not regulate it.
CPS I provides a way to form a (C=O)NH2 unit for urea by condensing bicarbonate and ammonia to form a carbamate, which contains a high-energy "motif" with respect to its hydrolysis product. (Note: This does not imply that the broken bond is high-energy, a misnomer found in many books.) Hence, the reaction is powered by 2 ATPs as an energy source. Urea is a molecule that "carries" both NH3/NH4+ and carbonate.
A reaction mechanism for carbamoyl phosphate synthase is shown in Figure \(\PageIndex{2}\).
Figure \(\PageIndex{2}\): Reaction mechanism for carbamoyl phosphate synthase
The two high-energy motif molecules (again with respect to their hydrolysis products) are protected from nonspecific hydrolysis as the mixed anhydride passes along a sequestered tunnel to a more distal phosphorylation site in the multimeric enzyme.
Figure \(\PageIndex{3}\) shows an interactive iCn3D model of the human carbamoyl phosphate synthetase I with bound ADP and N-acetyl-glutamate (5DOU).
Figure \(\PageIndex{3}\): Human carbamoyl phosphate synthetase I with bound ADP and N-acetyl-glutamate (5DOU). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...yV8fW3mW9idy6A
The model shows one chain of the multimer. The two ADPs and single N-acetylglutamate (NLG) are shown in spacefill and labeled. Additional bound ions are shown (unlabeled).
The protein exists in a monomer-dimer equilibrium mixture. Each monomer has 1 NAG and 2 ADPs in the two binding sites required for the two phosphorylation steps, as shown in the reaction above. The two phosphorylation sites are connected by a tunnel allowing passage of the mixed anhydride to the carbamate phosphorylation site. The NAG is an allosteric modifier as it is not bound near the phosphorylation sites. On binding to the apo form of the enzyme, it elicits a large conformation change, allowing a competent enzyme conformation with a complete tunnel to predominate.
An image of the tunnel for one monomer of the human carbamoyl phosphate synthetase I is shown in Figure \(\PageIndex{4}\).
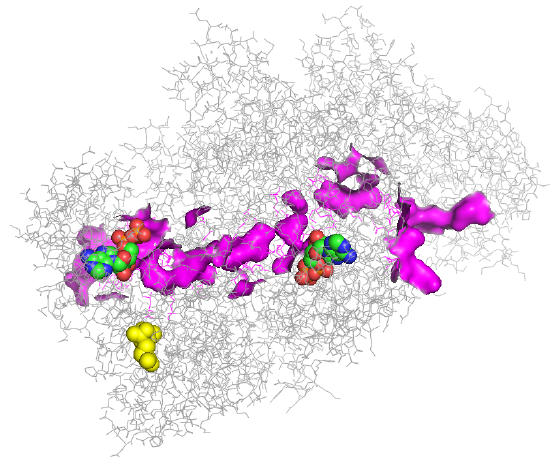
Figure \(\PageIndex{4}\): Tunnel for one monomer of the human carbamoyl phosphate synthetase I enzyme.
Two ADPs are shown below in spacefill with CPK color. The tunnel between the two ADP sites is highlighted in magenta. The yellow molecule is NAG, which again exerts its activation effect by binding to an allosteric site, effectively opening up the tunnel.
Figure \(\PageIndex{5}\) shows the enzyme monomer in a different orientation with a tunnel made using ChExVis: a tool for molecular channel extraction and visualization. The large red and green spheres show the surface openings.
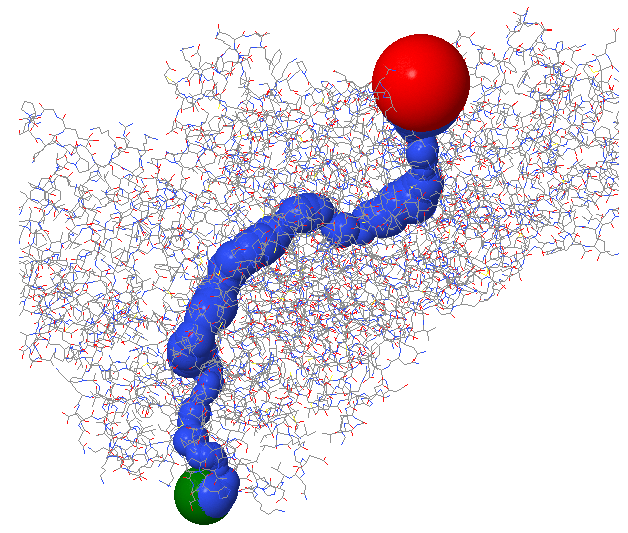
Figure \(\PageIndex{5}\): Second view of the tunnel in human carbamoyl phosphate synthetase I
The top panel in Figure \(\PageIndex{6}\) shows the radius in Angstroms of the tunnel going from the Red sphere to the Green sphere in the image above. The bottom panel measures the hydrophobicity (red)//hydrophilicity (blue) of the amino acids surrounding the tunnel.
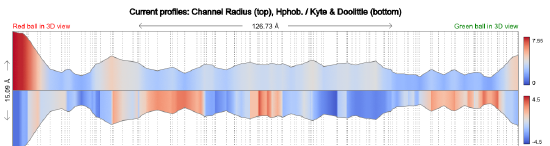
Figure \(\PageIndex{6}\): Tunnel size and polarity profile in human carbamoyl phosphate synthetase I
ASS1: ArginoSuccinate Synthase 1
Another ATP is cleaved to convert citrulline to arginosuccinate, as shown in Figure \(\PageIndex{7}\). The product could easily be cleaved in the next step to form the amino acid arginine.
Figure \(\PageIndex{7}\): ATP cleavage drives the conversion of citrulline to arginosuccinate
ASL: arginosuccinase lyase (aka arginosuccinase)
This enzyme catalyzes a beta-elimination reaction, which may proceed through an E2 or E1 mechanism. A general but very abbreviated mechanism showing a two-step (E1) elimination proceeding through a carbanion intermediate is shown in Figure \(\PageIndex{8}\) below.
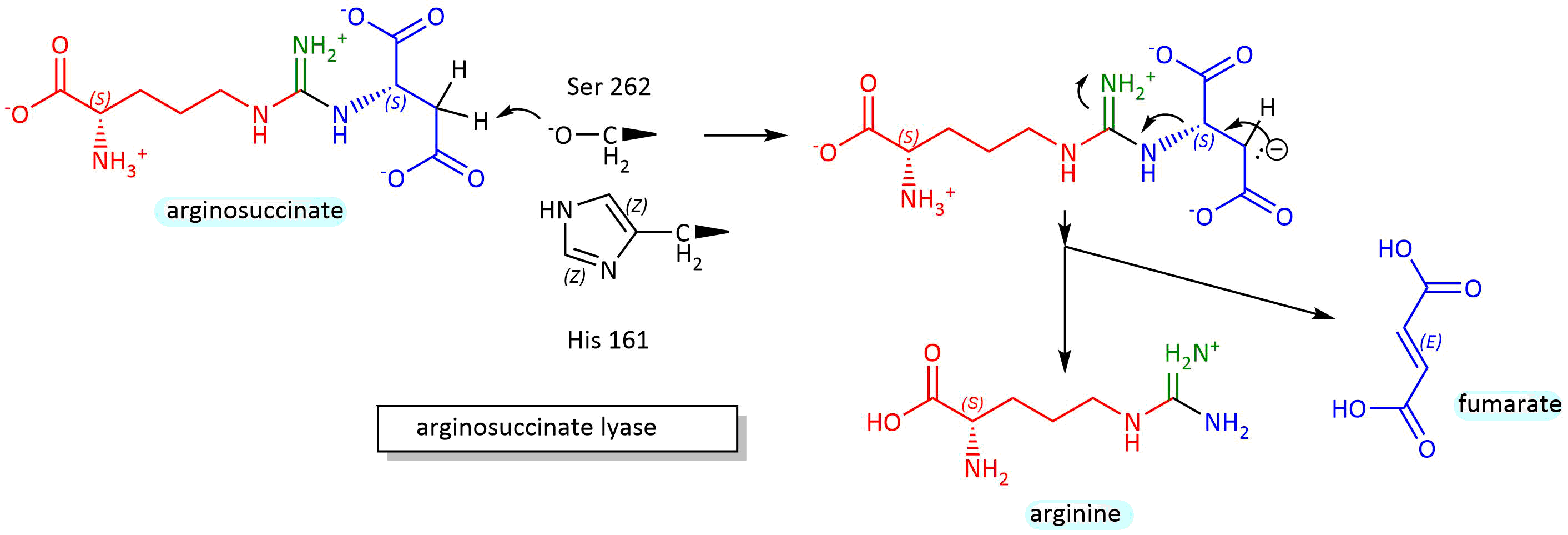
Figure \(\PageIndex{8}\): Mechanism for conversion of arginosuccinate to arginine and fumarate
Figure \(\PageIndex{9}\) shows an interactive iCn3D model of the tetrameric arginosuccinate lyase (ASL) from Mycobacterium tuberculosis (6IEN).
Figure \(\PageIndex{9}\): Tetrameric arginosuccinate lyase (ASL) from Mycobacterium tuberculosis (6IEN). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...32EuouGQ6xMVk6
Each monomer of the four monomers is shown in a different color. Three arginosuccinates (substrates) and fumarate, as well as arginine (products), are shown in spacefill with CPK colors and labeled. Each monomer has an N-, M-, and a C-domain. The four binding sites consist of residues from three monomers. The products are in the fourth site. Catalytic residues probably include a serine and a histidine acting as general acids/bases with a deprotonated lysine, making the serine a general base.
Ornithine Transcarbamoylase
The reaction mechanism is quite simple here. The carbamoyl phosphate is an activated electrophile in which a deprotonated amino group of ornithine attacks the carbonyl C to produce citrulline. The gene is found on the X chromosome in humans, so mutations in males lead to a "hyperammonemia coma" and death. Heterozygous females are either asymptomatic or have problems arising from defects in pyrimidine biosynthesis, which a low-protein diet can alleviate with the addition of arginine.
Arginase
There are two general variants of this enzyme. Arginase I is a cytosolic enzyme expressed in the liver and is involved in urea synthesis. Arginase II is a mitochondrial enzyme involved in arginine metabolism more generally. Arginase I converts arginine to ornithine and urea. The enzyme has 2 Mn ions in the active site. A hydroxide ion bridges the two Mn ions and acts as a nucleophile, attacking the arginine guanidino group and forming a tetrahedral intermediate. An active site aspartate (Asp 128) acts as a general acid, protonating the amino leaving group to form ornithine and urea. Water then re-adds to the Mn dinuclear cluster and ionizes, donating a proton to the solvent through an intermediary His 41 as it reforms the active enzyme.
The model below depicts the trimeric human arginase I (3KV2) in complex with the inhibitor N(omega)-hydroxy-nor-L-arginine, in which one of the guanidino nitrogens is replaced with an OH group. The two gray spheres are Mn ions.
Figure \(\PageIndex{10}\) shows an interactive iCn3D model of the trimeric human arginase I in complex with the inhibitor N(omega)-hydroxy-nor-L-arginine (3KV2).
Figure \(\PageIndex{10}\): Trimeric human arginase I in complex with the inhibitor N(omega)-hydroxy-nor-L-arginine (3KV2). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...QaAQpt5t2KKic6
Figure \(\PageIndex{11}\) show an active site water (#480) near the Mn ions.
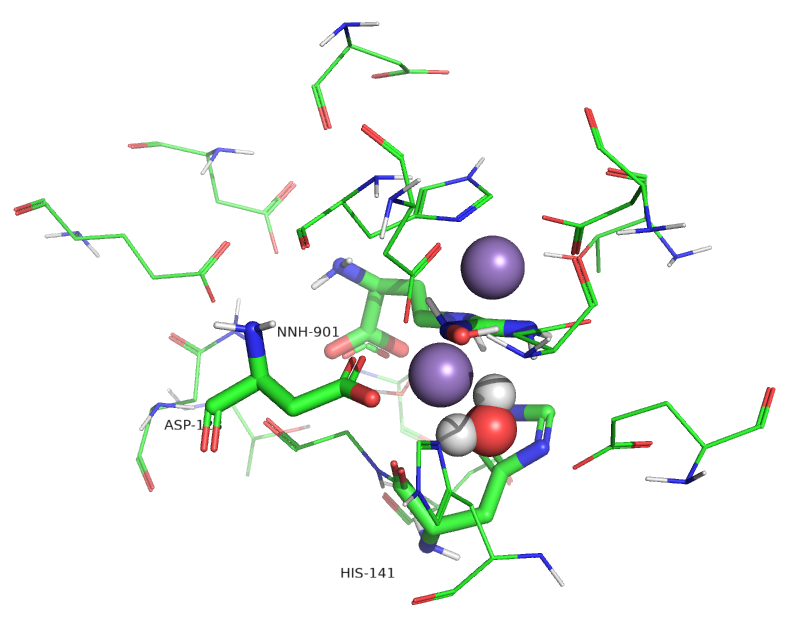
Figure \(\PageIndex{11}\): Active site of human arginase I with an active site water
Water molecules can act as ligands and bind transition-state metal ions through a coordinate covalent bond. The pKa of the bound water shifts to a lower value, making it more likely to deprotonate to form OH-, which is both a better base and nucleophile. Water 480 in the active site appears to form a coordinate covalent bond to the Mn ion, forming an active site hydroxide that attacks the guanidino group of arginine, leading to the formation of urea and ornithine.
Regulation
If excess amino acids/proteins are consumed, a change in gene expression can increase levels of the enzymes in the urea cycle. In the short term, the activity is regulated by the enzyme that provides access into the cycle, carbamoyl phosphate synthase, whose activity is regulated by N-acetylglutamic acid and which catalyzes the rate-limiting step. As shown above, the enzyme is functionally inactive in the absence of the allosteric regulator, NAG. The levels of NAG are determined by the enzyme NAG synthase, which is regulated by free arginine, the immediate precursor of urea in the cycle. Supplemental arginine is given to patients with urea cycle disorder, and its effect probably occurs through the regulation of NAG synthase.
The link between the urea cycle and the TCA cycle
Two major pathways you have explored, the TCA and urea cycles, are not linear but cyclic. What advantages do "circular" pathways offer over linear ones? Perhaps a comparison to economic pathways offers a clue. In a linear economy, raw materials are used to produce a product, which is eventually discarded as trash. Unfortunately, linear pathways dominate our world at a great cost to our environment. To reduce the inefficiencies in a linear economy, make it more viable, and decrease environmental damage and associated climate change, the world should move to a circular economy. A circular economy is characterized by what has been referred to as the 3Rs: reduce, reuse, and recycle. This saves resources and energy. These characteristics also apply to circular biochemical pathways, as reduced levels of metabolites are reused and recycled.
Given this, it should not be surprising that the urea and TCA cycles are linked, especially given that fumaric acid is a product of the urea cycle and a cyclic metabolite of the TCA cycle. In addition, aspartic acid is one transamination step away from oxaloacetate. The interconnections between the urea and TCA cycles are shown in Figure \(\PageIndex{12}\), along with two additions, the malate aspartate shuttle and the aspartate arginosuccinate shunt.
Figure \(\PageIndex{12}\): Urea and TCA cycles linked by the malate aspartate shuttle and the aspartate arginosuccinate shunt.
One of the nitrogens in urea comes from the amine of aspartate, which can be formed through the transamination of oxaloacetate from the TCA cycle to form aspartate. Since the TCA cycle is a cycle, an equivalent of one oxaloacetate must return to the cycle. It does so indirectly from fumarate, a TCA intermediate produced as a by-product of the urea cycle, but the fumarate is produced in the cytoplasm. There, it gets converted to malate, which can be transported into the mitochondria by a transport protein, mitochondrial 2-oxoglutarate/malate transporter, that is part of yet another cycle shown in the figure above, the aspartate arginosuccinate shunt. Fumarate is introduced into the shuttle through the aspartate-argininosuccinate shunt, as shown in the figure above. Once malate enters the mitochondrial matrix, it can be directly converted to oxaloacetate.
Note that these linked cycles require both mitochondrial and cytoplasmic forms of several enzymes and a balance between two key amino acids across the mitochondrial divide, glutamate and aspartate. One such key enzyme is aspartate aminotransferase (AST), also known as glutamate oxaloacetate transaminase (GOT), named for the reverse reaction of aspartate + α-ketoglutarate ↔ oxaloacetate + glutamate. As mentioned in the previous chapter section, this key enzyme can be detected in the blood if the liver is damaged, so its levels are routinely used in medical tests to assess impaired liver function.
Another key function of the malate-aspartate shuttle is to bring into the mitochondria reduced "equivalents" of NADH produced in the cytoplasm from glycolysis and fatty acid oxidation. There is no membrane transporter for NAD+ or NADH. Instead, cytoplasmic malate, produced by NADH's cytosolic reduction of oxaloacetate, can be transported into the matrix, which can reform mitochondrial NADH on reconversion to oxaloacetate. The NADH can then power ATP production through mitochondrial electron transport/oxidative phosphorylation pathways.
From an energetic perspective, four phosphoanhydride bonds in 3 ATP molecules are used to produce one urea molecule. This large energy expenditure is partially compensated for by ATP made from the "fumarate equivalents" that enter the mitochondria through the aspartate arginosuccinate and aspartate arginosuccinate shunts and the passage of NADH through oxidative phosphorylation.
Summary
The urea cycle is a central metabolic pathway that detoxifies excess ammonia produced during amino acid catabolism, converting it into urea—a water‐soluble, nontoxic molecule suitable for excretion. In the liver, ammonia is generated primarily via oxidative deamination, and excess nitrogen from a high-protein diet is ultimately processed through this cyclic pathway. The cycle uses energy, consuming three ATP molecules per urea molecule synthesized, a cost partially offset by the integration with other metabolic pathways.
Key Enzymes and Mechanisms:
The cycle involves several critical enzymes:
- Carbamoyl Phosphate Synthetase I (CPS I): Initiates the cycle by condensing bicarbonate with ammonia (using N-acetyl-glutamate as an allosteric activator) to form carbamoyl phosphate. The enzyme's structure features a substrate tunnel that channels high-energy intermediates between phosphorylation sites.
- Ornithine Transcarbamoylase (OTC): Catalyzes the reaction between carbamoyl phosphate and ornithine to form citrulline, highlighting the cycle’s regenerative nature.
- Argininosuccinate Synthase (ASS1) and Argininosuccinate Lyase (ASL): Work in tandem to combine citrulline with aspartate (which donates one of the nitrogen atoms) to form argininosuccinate, and then cleave it into arginine and fumarate. The reaction mechanisms here involve ATP cleavage and may proceed via elimination reactions.
- Arginase (ARG1): Converts arginine into urea and ornithine, the latter being recycled back into the cycle. The active site of arginase includes a dinuclear Mn center, which plays a crucial role in the hydrolytic mechanism.
Regulation and Metabolic Integration:
The urea cycle is tightly regulated at multiple levels:
- Allosteric Regulation: CPS I is dependent on N-acetyl-glutamate (NAG), whose levels are regulated by arginine, linking enzyme activity directly to nitrogen load.
- Gene Expression: Long-term adaptations to high protein intake involve changes in the expression of urea cycle enzymes.
- Connection to the TCA Cycle: Metabolic interconnections are significant. Fumarate produced in the cycle feeds into the TCA cycle, while aspartate—derived from oxaloacetate via transamination—links back to nitrogen metabolism. Additionally, shuttles like the malate-aspartate shuttle help transport reducing equivalents and maintain metabolic balance across cellular compartments.
Structural Insights:
Advances in structural biology have illuminated the architecture of key enzymes in the cycle. For example, interactive models of CPS I reveal how a conformational change induced by NAG binding opens a tunnel necessary for efficient substrate channeling, while studies on argininosuccinate lyase and arginase illustrate the multi-subunit organization and active site features that facilitate catalysis.
Clinical and Biochemical Significance:
Defects in any step of the urea cycle can result in severe metabolic disorders, such as hyperammonemia. Understanding the cycle's regulation and its integration with other metabolic pathways is essential for appreciating how disruptions in enzyme activity or localization may impact overall metabolic health. Moreover, the cyclic nature of the pathway provides a model for understanding the efficiency of metabolic recycling, drawing parallels to broader concepts such as circular economies.
Overall, the urea cycle is not only a key detoxification pathway but also an exemplary model of metabolic regulation, enzyme mechanism, and the integration of cellular pathways in response to dietary and physiological demands.




.png?revision=1&size=bestfit&width=381&height=276)
_from_Mycobacterium_tuberculosis_(6IEN).png?revision=1&size=bestfit&width=359&height=387)
-hydroxy-nor-L-arginine_(3KV2).png?revision=1&size=bestfit&width=325&height=331)