1.4: DNA Modifying Enzymes
- Page ID
- 18122
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Methylases
Just as the study of the bacterial restriction-modification system has provided a variety of specific endonucleases, there are also available a variety of specific DNA methylases.
- The recognition sequences of the methylases are the same as the associated endonucleases (e.g. EcoR1 methylase recognizes and methylates at the sequence "GAATTC").
- All methylases transfer the methyl group from S-adenosylmethionine (SAM) to a specific base in the recognition sequence, and SAM is a required component in the methylation reaction.
- Methylation of DNA usually has the effect of protecting the DNA from the related restriction endonuclease. However, there are methylases with minimal specificity. For example, Sss I methylase will methylate cytosine residues in the sequence 5' … CG … 3'. In this case, the methylated DNA will be protected from a wide variety of restriction endonucleases.
- Some restriction endonucleases will only cut DNA at their recognition sites if the DNA is methylated (e.g. Dpn I).
- Still other restriction endonucleases will cut both methylated and non-methylated DNA at their recognition sequences (e.g. BamH I).
dam and dcm Methylation
- The methylase encoded by the dam gene (dam methylase) transfers a methyl group from SAM to the N6 position of the adenine base in the sequence 5' … GATC … 3'.
- The methylase encoded by the dcm gene (dcm methylase) methylates the internal cytosine base, at the C5 position, in the sequences 5' … CCAGG … 3' and 5' … CCTGG … 3'.
- Almost all strains of E. coli commonly used in cloning have a dam+dcm+ genotype. The point here is not that we particularly want our DNA to be methylated, but that to make a dam-dcm- host someone has to mutate the bacteria and isolate the correct mutant. That apparently has not been done for a lot of bacterial strains. Probably because the dam and dcm methylation affects only a small subset of potential restriction endonucleases
DNA isolated from dam+dcm+ strains will not actually be cut by a modest subset of available restriction endonucleases:
|
|
|
|
|
|---|---|---|---|
| TGATCA |
|
|
|
| GATC |
|
|
|
| ATCGAT |
|
|
|
| TCTAGA |
|
|
|
| TCGA |
|
|
|
| GAAGA |
|
|
|
| GGTGA |
|
|
|
DNA may have to be prepared from E. coli strains which are dam-dcm- in order to be cut by these enzymes.
DNA Polymerases
A wide variety of polymerases have been characterized and are commercially available. All DNA polymerases share two general characteristics:
- They add nucleotides to the 3'-OH end of a primer
- The order of the nucleotides in the nascent polynucleotide is template directed
.png?revision=1&size=bestfit&width=580&height=226)
Figure 1.4.1: DNA Replication
In addition to the 5'->3' polymerase activity, polymerases can contain exonuclease activity. This exonuclease activity can proceed either in the 5'->3'direction, or in the 3'->5' direction.
- Exonuclease activity in the 3'->5' direction allows the polymerase to correct a mistake if it incorporates an incorrect nucleotide (so called "error correction activity"). It can also slowly degrade the 3' end of the primer.
- Exonuclease activity in the 5'->3' direction will allow it to degrade any other hybridized primer it may encounter. Without 5'->3' exonuclease activity, obstructing primers may or may not be physically deplaced, depending on the polymerase being used.
Different polymerases have differing error rates of misincorporation, and different rates of polymerization.
|
|
|
|
|
|
|
|---|---|---|---|---|---|
| 5'->3' exonuclease activity |
|
|
|||
| 3'->5' exonuclease activity |
|
|
|
|
|
| Error Rate (x10-6) |
|
|
|
|
|
| Strand Displacement |
|
||||
| Heat Inactivation |
|
|
|
|
Uses of polymerases
The various activities of the different polymerases lend them to a variety of applications. For example, restriction endonucleases can yield fragments of DNA with either 3' or 5' nucleotide "overhangs".
- In the case of 5' overhangs, the 5'->3' polymerase activity can fill these in to make blunt ends.
- In the case of 3' overhangs, the 3'->5' exonuclease activity present in some polymerases (especially T4 DNA polymerase) can "chew back" these ends to also make blunt-ended DNA fragments.
.png?revision=1&size=bestfit&width=839&height=412)
Figure 1.4.2: Polymerase activity
"Nick-translation"
This method is used to obtain highly radiolabeled single strand DNA fragments, which makes use of 5'->3' exonuclease activity present in some polymerases (E. coli DNA polymerase I, for example).
- In this method a DNA duplex of interest is "nicked" (i.e. one of the strands is cut; see DNAse I).
- Then DNA pol I is added along with radiolabeled nucleotides. The 5'->3' exonuclease activity chews away the 5' end at the "nick" site and the polymerase activity incorporates the radiolabeled nucleotides. The resulting polynucleotide will be highly radiolabeled and will hybridize to the DNA sequence of interest.
.png?revision=1&size=bestfit&width=799&height=195)
Figure 1.4.3: Nick-translation
- Thermostable polymerases have the ability to remain functional at temperature ranges where the DNA duplex will actually "melt" and become separated. This has allowed the development of the "Polymerase Chain Reaction" technique (PCR), which has had a profound impact on modern Biotechnology. We will discuss this method at a later date.
- The incorporation of dideoxy bases (i.e. no hydroxyl groups on either the 2' or 3' carbon of the ribose sugar) leads to termination of the polymerase reaction. This will be discussed in greater detail later. However, this chain termination by incorporation of dideoxynucleotides is the basis of the Sanger method of DNA sequencing, as well as therapies to try to inhibit viral replication.
Nucleases
Nuclease BAL-31
- This is an exonuclease (starts at the termini and works inward) which will degrade both 3' and 5' termini of double-stranded DNA. It will not make internal cleavages ("nicks"), however, it will degrade the ends of DNA at existing internal "nicks" (which create both 3' and 5' termini).
- The degradation of termini is not coordinated, meaning that the product is not 100% blunt-ended (even though the original duplex may have been blunt ended).
- Such "ragged" ends can be made blunt by filling in and chewing back by a suitable polymerase (e.g. T4 DNA polymerase). The unit definition of 1 unit is the amount of enzyme required to remove 200 base pairs from each end of duplex DNA in 10 minutes at 30 °C.
.png?revision=1&size=bestfit&width=725&height=230)
Figure 1.4.4: Nuclease BAL-31 activity
Exonuclease III
- Catalyzes the stepwise removal of nucleotides from the 3' hydroxyl termini of duplex DNA.
- The enzyme will attack the 3' hydroxyl at duplex DNA with blunt ends, with 5' overhangs, or with internal "nicks".
- Since duplex DNA is required, the enzyme will not digest the 3' end of duplex DNA where the termini are 3' overhangs.
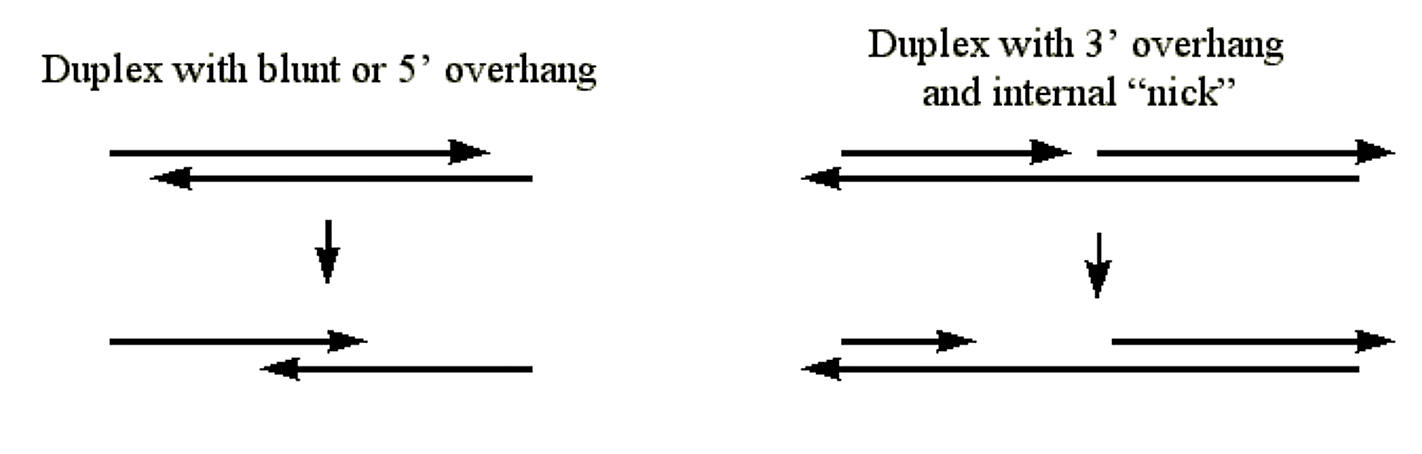.png?revision=1&size=bestfit&width=720&height=230)
Figure 1.4.5: Exonuclease III Activity
Mung Bean Nuclease (isolated from mung bean sprouts)
- A single strand specific DNA and RNA endonuclease which will degrade single strand extensions from the ends of DNA and RNA leaving blunt ends.
- The single strand extensions can be either 5' or 3' extensions - both are removed and a blunt duplex is left.
.png?revision=1&size=bestfit&width=728&height=288)
Figure 1.4.6: Mung Bean Nuclease activity
Deoxyribonuclease I (DNAse I) from Bovine pancrease
- This enzyme hydrolyzes duplex or single DNA strands preferentially at the phosphodiester bonds 5' to pyrimidine nucleotides
- In the presence of Mg2+ ion, DNAse I attacks each strand independently and produces nicks in a random fashion (useful for nick-translation)
- In the presence of Mn2+ ion DNAse I cleaves both strands of DNA at approximately the same position (but leaving "ragged" ends)
Ligases
- Ligases catalyze the formation of a phosphodiester bond between juxtaposed 5' phosphate and 3' hydroxyl termini of nucleotides (potentially RNA or DNA depending on the ligase).
- In a sense, they are the opposite of restriction endonucleases, but they do not appear to be influenced by the local sequence, per se.
- Ligases require either rATP or NAD+ as a cofactor, and this contrasts with restriction endonucleases.
The following are different types of ligases and their characteristics.
T4 DNA ligase
- Isolated from bacteriophage T4.
- Will ligate the ends of duplex DNA or RNA.
- This enzyme will join blunt-end termini as well as ends with cohesive (complementary) overhanging ends (either 3' or 5' complementary overhangs).
- This enzyme will also repair single stranded nicks in duplex DNA, RNA or DNA/RNA duplexes. Requires ATP as a cofactor.
Taq DNA ligase
- This ligase will catalyze a phosphodiester bond between two adjacent oligonucleotides which are hybridized to a complementary DNA strand:
.png?revision=1&size=bestfit&width=291&height=102)
Figure 1.4.7: Taq DNA ligase activity
- The ligation is efficient only if the oligonucleotides hybridize perfectly with the template strand.
- The enzyme is active at relatively high temperatures (45 - 65 °C). Requires NAD+ as a cofactor.
T4 RNA ligase
- Will catalyze formation of a phosphodiester bond between RNA/RNA oligonucleotides, RNA/DNA oligonucleotides, or DNA/DNA oligonucleotides.
- Requires ATP as a cofactor.
- This enzyme does not require a template strand.
T4 RNA ligase can be used for a variety of purposes including constructing RNA/DNA hybrid molecules.
.png?revision=1&size=bestfit&width=548&height=224)
Figure 1.4.8: T4 RNA ligase activity
DNA ligase (E. coli)
- Will catalyze a phosphodiester bond between duplex DNA containing cohesive ends.
- It will not efficiently ligate blund ended fragments.
- Requires NAD+ as a cofactor.
.png?revision=1&size=bestfit&width=552&height=158)
Figure 1.4.9: DNA ligase (E. Coli) activity
T4 polynucleotide kinase
- Catalyzes the transfer and exchange of a phosphate group from the g position of rATP (adenine ribose triphosphate nucleotide) to the 5' hydroxyl terminus of double stranded and single stranded DNA or RNA, and nucleoside 3' monophosphates.
- The enzyme will also remove 3' phosphoryl groups.
- Oligonucleotides which are obtained from automated synthesizers lack a 5' phosphate group, and thus, cannot be ligated to other polynucleotides.
T4 polynucleotide kinase can be used to phosphorylate the 5' end of such polynucleotides:
.png?revision=1&size=bestfit&width=580&height=221)
Figure 1.4.10: T4 polynucleotide kinase activity
Calf intestinal phosphatase (CIP)
- Catalyzes the removal of 5' phosphate groups from RNA, DNA and ribo- and deoxyribo- nucleoside triphosphates (e.g. ATP, rATP).
- CIP treated duplex DNA cannot self ligate.
- Hemi-phosphorylated duplexes will be ligated on one strand (the phosphorylated strand) and remain "nicked" on the other.
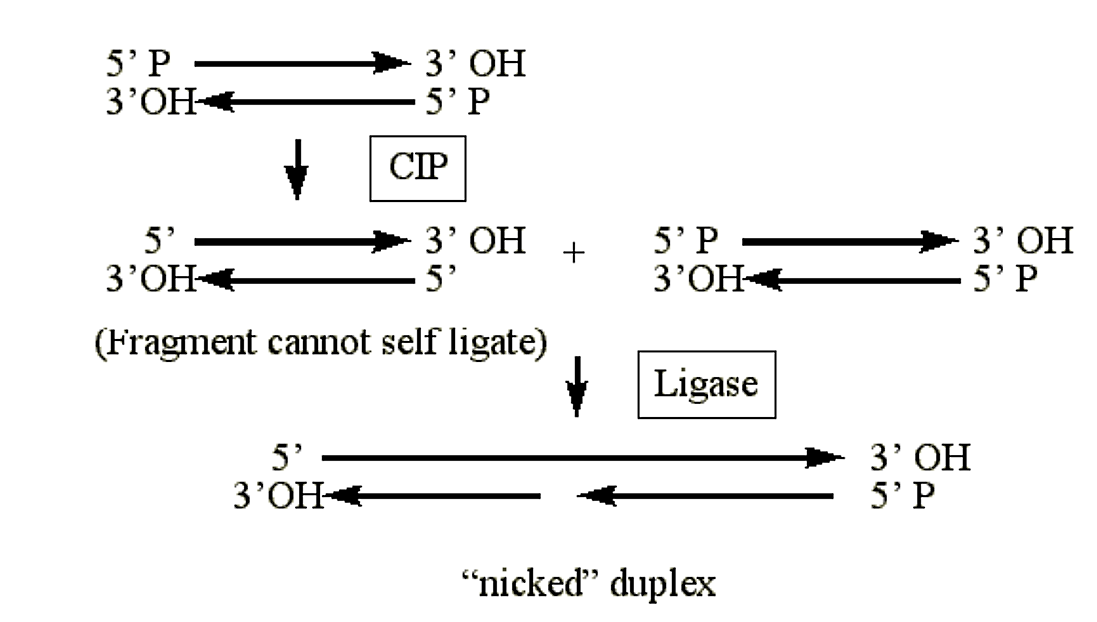.png?revision=1&size=bestfit&width=614&height=348)
Figure 1.4.11: CIP activity


