4.6: Intrinsically Disordered Proteins
- Page ID
- 62323
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)-
Define and Characterize Intrinsic Disorder:
- Explain what intrinsically disordered proteins (IDPs) and intrinsically disordered regions (IDRs) are, and describe how their amino acid composition (enrichment in polar and charged residues with low hydrophobicity) distinguishes them from ordered proteins.
- Interpret experimental data (e.g., from X-ray crystallography, NMR, CD spectroscopy, and computational predictions such as AlphaFold confidence scores) that indicate the presence of disorder in proteins.
-
Understand the Functional Role of Protein Disorder:
- Discuss how the flexible, dynamic nature of IDPs allows them to bind multiple partners and participate in cell signaling, regulation, and transcription.
- Describe the concept of coupled folding and binding, and explain how disorder enables a protein to adopt more ordered structures upon ligand binding.
- Evaluate the idea of the "Protein Trinity," which posits that proteins exist in multiple native states (ordered, collapsed, and extended) that contribute to their biological functions.
-
Examine the Biological and Regulatory Implications of Disorder:
- Analyze how intrinsic disorder affects protein turnover, aggregation propensity, and post-translational modifications.
- Discuss the significance of IDPs in disease contexts, including their roles in neurodegenerative disorders (e.g., tauopathies and alpha-synuclein aggregation) and in the formation of dynamic complexes.
-
Explore Metamorphic Proteins:
- Define metamorphic proteins and distinguish them from both fully ordered proteins and prion proteins by discussing their reversible, alternative conformations.
- Describe how metamorphic proteins, such as lymphotactin (XCL1) and Mad2, can switch conformations in response to subtle changes (e.g., in pH, redox state, or binding interactions) without complete unfolding.
- Evaluate the evolutionary and functional significance of proteins that can adopt multiple, interconvertible conformations.
-
Apply Computational and Experimental Approaches:
- Illustrate how molecular dynamics simulations and AI-based prediction tools (e.g., ALBATROSS, IDRLab) are used to study the conformational ensembles of disordered proteins.
- Interpret interactive structural models (e.g., using iCn3D) to visualize examples of IDPs (such as SIC1, tau, and alpha-synuclein) and metamorphic proteins, and relate these structures to their functional outcomes.
-
Connect Sequence, Structure, and Function in Disorder:
- Explain how synonymous mutations can influence protein folding and structure, particularly for membrane proteins, by affecting the kinetics of translation and cotranslational folding.
- Discuss how the lack of a stable hydrophobic core in disordered regions underlies their dynamic behavior and functional versatility in the cell.
These learning goals provide a roadmap for understanding the diverse roles of protein disorder and metamorphism in cellular function and regulation, bridging concepts from structural biochemistry, molecular dynamics, and protein evolution.
Intrinsically Disordered Proteins (IDPs) and Metamorphic Proteins
Many examples of proteins that are partially or completely disordered yet retain biological function have been identified. At first glance, this might appear unexpected since how could such a protein bind its natural ligand with specificity and selectivity to express its function? Of course, one could postulate that ligand binding would induce conformational changes necessary for function (such as catalysis), in an extreme example of an induced fit rather than a "lock-and-key" fit. Decades ago, Linus Pauling predicted that antibodies, proteins that recognize foreign molecules (antigens), would bind loosely to the antigen, then undergo a conformational change to form a more complementary, tighter fit. This was the easiest way to allow a finite number of protein antibodies to bind to seemingly endless numbers of foreign molecules. This is indeed one way antibodies can recognize foreign antigens. Antibodies that bind to antigens with high affinity and, hence, high specificity are more likely to bind through a lock-and-key fit. (Pauling, however, didn't know that the genes that encode the protein chains in antibodies are differentially spliced and subjected to enhanced mutational rates, allowing the generation of incredible antibody diversity from a limited set of genes.)
Intrinsically Disordered Proteins (IDPs)
It's been estimated that over half of all native proteins have regions (greater than 30 amino acids) that are disordered, and upwards of 20% of proteins are completely disordered. Regions of disorder are enriched in polar and charged side chains, since these might be expected to assume many available conformations in aqueous solutions compared to sequences enriched in hydrophobic side chains, which would probably collapse into a compact core stabilized by the hydrophobic effect. Mutations in the disordered regions tend to preserve the disordered region, suggesting that the disordered region is advantageous for "future" function. In addition, mutations that convert a noncoding sequence into a coding one invariably yield disordered protein sequences. Disordered proteins tend to have regulatory properties and bind multiple ligands, whereas ordered proteins are involved in highly specific ligand binding necessary for catalysis and transport. The intracellular concentration of disordered proteins has also been shown to be lower than that of ordered proteins, possibly to prevent inappropriate binding interactions mediated by hydrophobic interactions, for example. Processes to accomplish this include more rapid mRNA and protein degradation and slower translation of mRNA for disordered proteins. For a similar reason, misfolded proteins are also targeted for degradation. Figure \(\PageIndex{1}\) shows characteristics of intrinsically disordered proteins.
Panel A shows the mean net charge vs the mean hydrophobicity for 275 folded and 91 natively unfolded proteins. Panel B shows the relative amino acid composition of globular (ordered) proteins compared to regions of disorder greater than 10 amino acids in disordered proteins. The two grey bars were obtained using two software versions to analyze the proteins. Again, the graph shows the enrichment of hydrophilic amino acids in disordered proteins.
Many experimental methods can detect disordered regions in proteins. Such regions are not well resolved in X-ray crystal structures (they have high B factors). NMR solution structures would show multiple, distinct conformations. CD spectroscopy likewise would show an ill-defined secondary structure. In addition, solution measurements of size (light scattering, centrifugation) would show more significant size distributions for a given protein.
What types of proteins contain disorder? The above experimental and new computational methods have been developed to classify proteins according to their degree of disorder. There appear to be more IDPs in eukaryotes than in archaea and prokaryotes. Many IDPs are involved in cell signaling processes (when external molecules signal cells to respond by proliferating, differentiating, dying, etc). Most appear to reside in the nucleus. The largest percentage of known IDPs bind to other proteins and DNA. These results suggest that IDPs are essential to protein function and probably confer significant advantages to eukaryotic cells. Multiple functions can be elicited from the interaction of a single IEP (derived from a single gene) with different protein-binding partners. This would greatly extend the effective genome size in humans from around 20,000 protein-encoding genes with specified functions to many more. This doesn't even consider the increase in functionalities derived from post-translational chemical modifications.
Protein structure is fluid and complex, and our simple notions and words to denote proteins as either native or denatured are misguided and constrain our ideas about how protein structure elicits biological function. For example, what does the word "native" mean if proteins exist in multiple states simultaneously, in vivo and in vitro? Dunker et al. (2001) coined the concept of the "Protein Trinity" to move beyond the notion that a single protein folds to a single state, which elicits a single function. Instead, each of the states in the "trinity", the ordered, collapsed (or molten globule) and extended (random coil), coexist in the cell, as shown in Figure \(\PageIndex{2}\). Characteristics of Intrinsically Disordered Proteins. Hence, all can be considered "native" and all contribute to the cell's function. A single IDP could bind to multiple protein partners, yielding distinct final structures and functions. IDPs would also be more accessible and hence more susceptible to proteolysis, providing a simple mechanism to control their concentrations —an important way to regulate their biological activity. Their propensity to undergo post-translational chemical modification would lead to new biological regulation types.
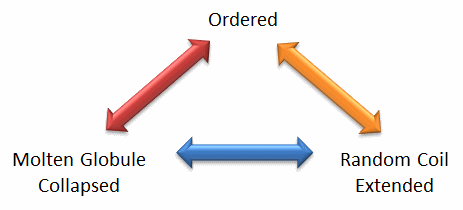
These ideas have profound ramifications for our understanding of cellular phenotype expression. In addition, a whole new world of drug targets is available, as drugs that modulate transitions between ordered, collapsed, and extended protein states are being discovered. Likewise, side effects of drugs might be understood by investigating their effects on these transitions in IDPs that were not initially targeted for analysis. Several web databases, including PONDR - Predictor of Naturally Occurring Disorder and Database of Protein Disorder, are available.
IDPs span a spectrum from fully unstructured to partially structured, including random coils, (pre-)molten globules, and large multi-domain proteins connected by flexible linkers. They constitute one of the main types of protein (alongside globular, fibrous, and membrane proteins). Figure \(\PageIndex{3}\) shows the conformational flexibility in SUMO-1 protein (PDB:1a5r), which is a composite of 10 NMR structures. The central part shows a relatively ordered structure. Conversely, the N- and C-terminal regions (left and right, respectively) show ‘intrinsic disorder’.
Figure \(\PageIndex{3}\): Conformational flexibility in SUMO-1 protein (1a5r) showing intrinsically disordered regions
It's interesting to explore the history of our understanding of IDPs. In the 1930s -1950s, the first protein structures were solved by protein crystallography. These early structures suggested that a fixed three-dimensional structure might be generally required to mediate the biological functions of proteins. When stating that proteins have just one uniquely defined configuration, Mirsky and Pauling did not recognize that Fisher's work would have supported their thesis with his 'Lock and Key' model (1894). These publications solidified the central dogma of molecular biology: the sequence determines the structure, which, in turn, determines the function of proteins. In 1950, Karush wrote about 'Configurational Adaptability,' contradicting all the assumptions and research of the 19th century. He was convinced that proteins have more than one configuration at the same energy level and can choose one when binding to other substrates. In the 1960s, Levinthal's paradox suggested that the systematic conformational search of a long polypeptide is unlikely to yield a single folded protein structure on biologically relevant timescales (i.e., seconds to minutes). Curiously, for many (small) proteins or protein domains, in vitro refolding can be rapid and efficient. As stated in Anfinsen's Dogma from 1973, the fixed 3D structure of these proteins is uniquely encoded in their primary structure (the amino acid sequence), is kinetically accessible and stable under a range of (near) physiological conditions, and can, therefore, be considered as the native state of such "ordered" proteins.
During the subsequent decades, however, many large protein regions could not be assigned in X-ray datasets, indicating that they occupy multiple positions, which average out in electron density maps. The lack of fixed, unique positions relative to the crystal lattice suggested that these regions were "disordered". Nuclear magnetic resonance spectroscopy of proteins also demonstrated the presence of large flexible linkers and termini in many solved structural ensembles. It is generally accepted that proteins exist as ensembles of similar structures, with some regions more constrained than others.
Some people differentiate a particular type of IDP called Intrinsically Unstructured Proteins (IUPs), which occupy the end of this flexibility spectrum. In contrast, IDPs also include proteins with a strong local structural tendency or flexible multidomain assemblies. These highly dynamic disordered regions of proteins have subsequently been linked to functionally important phenomena such as allosteric regulation and enzyme catalysis.
Many disordered proteins have their binding affinity for receptors regulated by post-translational modifications. Hence, it has been proposed that the flexibility of disordered proteins facilitates the attainment of the conformational requirements for binding their modifying enzymes and receptors. Intrinsic disorder is particularly common in proteins involved in cell signaling, transcription, and chromatin remodeling. Here are some types or characteristics of IDPs.
Flexible linkers
Disordered regions are often found as flexible linkers or loops connecting domains. Linker sequences vary greatly in length but are typically rich in polar uncharged amino acids. Flexible linkers allow the connecting domains to twist and rotate freely, recruiting their binding partners through protein domain dynamics. They also allow their binding partners to induce larger-scale conformational changes by long-range allostery.
Linear motifs
Linear motifs are short, disordered segments of proteins that mediate functional interactions with other proteins or biomolecules (RNA, DNA, sugars, etc.). Many roles of linear motifs are associated with cell regulation, for instance, in controlling cell shape, subcellular localization of individual proteins, and regulated protein turnover. Post-translational modifications, such as phosphorylation, often tune the affinity (not infrequently by several orders of magnitude) of individual linear motifs for specific interactions. Unlike globular proteins, IDPs lack preformed active sites. Nevertheless, in 80% of IDPs (~3 dozen) subjected to detailed structural characterization by NMR, linear motifs termed PreSMos (pre-structured motifs) —transient secondary structural elements primed for target recognition —are identified. In several cases, it has been demonstrated that these transient structures become full and stable secondary structures, e.g., helices, upon target binding. Hence, PreSMos are the putative active sites in IDPs.
Coupled folding and binding
Many unstructured proteins transition to more ordered states upon binding to their targets. Coupled folding and binding may be local, involving only a few interacting residues, or global, involving an entire protein domain. It was recently shown that coupled folding and binding allow the burial of a large surface area, which would be possible only for fully structured proteins if they were much larger. Moreover, certain disordered regions might serve as "molecular switches" that regulate biological functions by adopting ordered conformations upon binding small molecules, nucleic acids, or ions.
Disorder in the bound state (fuzzy complexes)
Intrinsically disordered proteins can retain their conformational freedom even when they bind specifically to other proteins. The structural disorder in the bound state can be static or dynamic. Structural multiplicity is required for function in fuzzy complexes, and manipulating the bound-disordered region alters activity. The conformational ensemble of the complex is modulated via post-translational modifications or protein interactions. The specificity of DNA-binding proteins often depends on the length of fuzzy regions, which vary through alternative splicing. Intrinsically disordered proteins adopt many different structures in vivo in response to cellular conditions, forming a structural or conformational ensemble.
Therefore, their structures are strongly function-related. However, only a few proteins are fully disordered in their native state. The disorder is mainly found in intrinsically disordered regions (IDRs) within an otherwise well-structured protein. Therefore, the term intrinsically disordered protein (IDP) includes proteins containing IDRs and fully disordered proteins.
The existence and kind of protein disorder are encoded in its amino acid sequence. As described above, IDPs are characterized by a low content of bulky hydrophobic amino acids and a high proportion of polar and charged amino acids, resulting in low hydrophobicity. This property leads to good interactions with water. Furthermore, high net charges promote disorder due to electrostatic repulsion between similarly charged residues. Thus, disordered sequences cannot sufficiently bury a hydrophobic core to fold into stable globular proteins. In some cases, hydrophobic clusters in disordered sequences provide clues to identify regions that undergo coupled folding and binding (see biological roles).
Many disordered proteins lack regular secondary structure. These regions can be considered more flexible than structured loops. While the latter are rigid and contain only one set of Ramachandran angles, IDPs involve multiple sets of angles. The term flexibility is also used for well-structured proteins, but describes a different phenomenon in the context of disordered proteins. Flexibility in structured proteins occurs around an equilibrium state, whereas it doesn't in IDPs. Many disordered proteins also contain low-complexity sequences, i.e., sequences over-representing a few residues. While low-complexity sequences are a strong indication of disorder, the reverse is not necessarily true. That is, not all disordered proteins have low-complexity sequences. Disordered proteins have low predicted secondary structure content.
Recent Updates: October - December, 2024
One way to determine the extent of disorder of a given protein is to examine the certainty in the predicted AlphaFold structure of the protein. Two examples of proteins that are largely disordered in AlphaFold models are shown below. The legend below shows the color corresponding to the confidence level of the predicted structure along the sequence.

Figure \(\PageIndex{4}\) shows interactive iCn3D model of AlphaFold predicted structure of the yeast SIC1 protein (P38634)

Figure \(\PageIndex{4}\): AlphaFold predicted the structure of the yeast SIC1 protein (P38634). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...rJsptsoszZGm29
The SIC1 is a cyclin-dependent kinase (CDK) inhibitor. It interacts with Cdc4, a subunit of ubiquitin ligase, which degrades SIC1. This regulates the G1-to-S cell cycle transition in budding yeast. SIC1 has nine sites for post-translational phosphorylation. Phosphorylation of individual sites enables this IDP to bind the active site of Cdc4. Especially important are phosphorylation of threonines 5, 33, and 45 and serines 69, 76, and 80.
Figure \(\PageIndex{5}\) shows interactive iCn3D model of the interactions of a phosphorylated pSIC1 peptide with key residues in Cdc4 from a ScSkp1-ScCdc4-pSic1 peptide complex (3V7D)
Figure \(\PageIndex{5}\): Interactions of a phosphorylated pSIC1 peptide with key residues in Cdc4 from a ScSkp1-ScCdc4-pSic1 peptide complex (3V7D). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...PZmqrqwmQyyJJ6
The pSIC1 peptide is shown with a cyan backbone. Ser 76 and Ser 80 are phosphorylated. Note the salt bridges (ion-ion interactions) with arginine side chains in the active site of Cdc4. Figure 4 in this paper shows a model of the interactions between phosphorylated SIC1 and Cdc4. Phosphorylation appears to cause only local, not global, ordering of the SIC1 protein. This local ordering provides sufficient interactions between SIC1 and Cdc4.
The second protein we'll discuss is the human microtubule-associated protein tau.
Figure \(\PageIndex{6}\) shows interactive iCn3D model of the AlphaFold predicted structure of the human microtubule-associated protein tau (P10636).
Figure \(\PageIndex{6}\): AlphaFold predicted structure of the human microtubule-associated protein tau (P10636). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/icn3d/share.html?6gs41N9MEbziWuJe8
This protein is almost entirely disordered. It promotes tubulin association into microtubules (as described in Chapter 5.5: Protein Interactions Modulated by Chemical Energy- Actin, Myosin, and Molecular Motors). It's especially important in neurons for axonal transport.
Figure \(\PageIndex{7}\) shows interactive iCn3D model of the cryoEM structure of human tau-tubulin (7PQC).
Figure \(\PageIndex{7}\): CryoEM structure of human tau-tubulin (7PQC). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/icn3d/share.html?nFiZfGoWZuSzDdT6A
This is quite an amazing structure! A microtubule consists of multiple copies of a dimer of two monomeric tubulins, alpha and beta. The tubulin monomers are shown in colored cartoons in the model above. Tau is shown in gray as the elongated spacefill structure. It consists of the tau-F isoform's microtubule-binding region (amino acids 202-395), which has 441 residues. Tau is largely still disordered even in the bound state! There are regions of closer noncovalent interactions with the microtubule. Key interacting residues in Tau are labeled and shown in red spacefill.
Tau has four highly homologous repeats R1–R4. Figure \(\PageIndex{8}\) shows electron microscopy (EM) models of Tau/MTs interaction showing two of the repeats, R1 and R2. Many of the interactions are salt bridges.
Figure \(\PageIndex{8}\): Electron microscopy (EM) models of Tau/MTs interaction. Barbier P, Zejneli O, Martinho M, Lasorsa A, Belle V, Smet-Nocca C, Tsvetkov PO, Devred F, and Landrieu I (2019). Role of Tau as a Microtubule-Associated Protein: Structural and Functional Aspects. Front. Aging Neurosci. 11:204. doi: 10.3389/fnagi.2019.00204 Creative Commons Attribution License (CC BY).
The α-tubulin subunit is represented as a blue cube, and the β-tubulin subunit as a brown cube. H12 C-terminal helix is schematized; the C-terminal tail prolongs the H12 helix. Models based on cryo-EM coupled to Rosetta modeling: contacts of the R1 repeat with the α-tubulin subunit at the inter-dimer interface: S258 and S262 of R1 make hydrogen bonds with E434; K259 of R1 interacts with an acidic patch formed by E420, E423, and D424; I260 of R1 is in a hydrophobic pocket formed by residues I265, V435 and Y262; K267 of R1 is in contact with the acidic C-terminal tail. Additional contacts for the R2 repeat with the β-tubulin subunit at the intra-dimer interface: K274 of R2 interacts with an acidic patch formed by D427 and S423; K281 of R2 is in contact with the acidic C-terminal tail of the β-tubulin subunit. The PHF6* peptide (highlighted green) is close to this tail and localizes at the intra-dimer interface. Additional contacts for the R2 repeat with the α-tubulin subunit: K294 and K298 are in contact with the acidic C-terminal tail of the α-tubulin subunit. Finally, H299 of R2 is buried in a cleft formed by residues F395 and F399 of the β-tubulin subunit.
Tau also interacts with RNA and DNA in the nucleus and at cell membranes. It self-associates to form aggregates associated with Alzheimer's and other neurodegenerative diseases (collectively called tauopathies). The latter is described in Chapter 4.10: Protein Aggregates - Amyloids, Prions and Intracellular Granules. Many post-translational modifications, including phosphorylation, ubiquitination, glycosylation, nitration, acetylation, and limited proteolysis, regulate tau's structure and activity. As above, phosphorylation plays a key role in regulating tau activity. Phosphorylated and acetylated tau have a lower affinity for the microtubules, which dysregulates their binding. The protein in aggregates is hyperphosphorylated.
Repeats 1-4 have both weak and strong interaction motifs. One strong site is the SK(I/C)GS motif. Phosphorylation within this motif is associated with Alzheimer's Disease. Experimental evidence implicates S235, S241, S262, S324, S356, K259, K311, K340 and K353 as key interaction residues. These are shown in Figure 7 above as labeled red spacefill amino acids in tau (gray) and are in close contact with the microtubule. The lysine interactions are modified and weakened by acetylation.
AI and machine learning have been deployed to analyze sequence data to identify intrinsically disordered regions (IDRs) in protein sequences. One such program is ALBATROSS, A deep-learning Approach for predicting the properties Of disordered proteins. It can predict conformational "ensembles" of disordered proteins, as disordered regions would be expected to have multiple conformations. It is available through Google Colaboratory. Another is IDRLab, which allows molecular dynamics simulations of intrinsically disordered proteins through Google Colab.
Figure \(\PageIndex{9}\) below shows a simplified molecular dynamics simulation of the intrinsically disordered protein human alpha-synuclein (Uniprot ID P37840). (Molecular dynamics were discussed in Chapter 3.4: Analyses of Protein Structure.) α-Synuclein (140 amino acids, MW 14,460) is expressed in the brain and presynaptic terminals in the central nervous system, but also in more distal neurons, and is involved in the regulation of neurotransmitter release and in the synaptic vesicles that hold them. Its aggregation is a cause or consequence of Parkinson's Disease and Lewy Body Dementia (see more details in Chapter 4.10: Protein Aggregates - Amyloids, Prions and Intracellular Granules).

Figure \(\PageIndex{9}\): Simplified molecular dynamics simulation of the intrinsically disordered protein human alpha-synuclein (Uniprot ID P37840). G. Tesei, A. I. Trolle, N. Jonsson, J. Betz, F. Pesce, K. E. Johansson, K. Lindorff-Larsen. Conformational ensembles of the human intrinsically disordered proteome: Bridging chain compaction with function and sequence conservation bioRxiv 2023 2023.05.08.539815 DOI: https://doi.org/10.1101/2023.05.08.539815
This Colab notebook enables running molecular dynamics (MD) simulations of intrinsically disordered proteins (IDPs) and protein regions (IDRs) and to study their conformational ensembles. MD simulations employ the coarse-grained force field CALVADOS2, in which each residue is mapped to a single bead and modeled with a stickiness parameter and electrostatics.
Note the extreme range of conformations of α-Synuclein, allowing this protein to interact with many possible protein targets and also itself when it forms aggregates. Compare it to a one-nanosecond molecular dynamics simulation of two proteins of similar molecular weight shown below in Figure \(\PageIndex{10}\). On the left, the hen egg white lysosome (2VB1) has four intramolecular disulfide bonds. The eight cysteines involved in the disulfide bonds are shown in spacefill. On the right is sperm whale myoglobin with the heme removed before the simulation (1A6M). This protein does not have disulfide bonds. Note that the lysozyme is dynamically more constrained by the 4 S-S bonds, making the whole structure slightly more rigid.
|
2VB1: Hen Egg White Lysozyme (4 S-S bonds) |
1A6M: Myoglobin (apo, no S-S bonds) |
Figure \(\PageIndex{10}\): 1-nanosecond simulations were performed on Gromacs using Neuroapp. The input PDB files for the simulations were first processed using Charmm-GUI. The output simulation files (input_protein and md_center.xtc from the output folder) from the Gromacs simulation were opened in Pymol and processed to make the animations shown above (File, Export Movie, as png. then animated gif in APS). Abraham, J. M. et al., GROMACS: High-performance molecular simulations through multi-level parallelism from laptops to supercomputers, https://www.sciencedirect.com/, September 2015, https://doi.org/10.1016/j.softx.2015.06.001. Neurosnap Inc. - Computational Biology Platform for Research. Wilmington, DE, 2022. https://neurosnap.ai/.
Click the link below for more information.
- The tick saliva glycine-rich protein!
-
As described above, IDPs are prone to aggregate, with some aggregates being quite amorphous. One example of such an aggregate is the "cement cone" formed by a tick that allows it to stick to its host. About 20% of the aggregate consists of the tick saliva protein glycine-rich protein (GRP). Noncovalent interactions hold together the aggregate/cement. These processes are illustrated in Figure \(\PageIndex{x}\), along with the amino acid characteristics of the protein.
Figure \(\PageIndex{x}\): Tick Glycine Rich Protein Secretion. Ganar, K.A., Nandy, M., Turbina, P. et al. Phase separation and aging of glycine-rich protein from tick adhesive. Nat. Chem. (2024). https://doi.org/10.1038/s41557-024-01686-8. Creative Commons Attribution 4.0 International License. http://creativecommons.org/licenses/by/4.0/.
Panel a: Schematic overview showing the consequences of a tick bite. The tick inserts its hypostome into the host epidermis and secretes a protein-rich saliva abundant in GRPs. The saliva undergoes a liquid-to-solid transition, forming a hard cement cone that allows the tick to feed on the host for several days and facilitates pathogen transmission (the shown ‘bull’s-eye’ rash is typical in the case of Borrelia infection, causing Lyme borreliosis).
Panel b: Amino-acid composition of tick-GRP77 shows a high proportion of non-polar amino acids (44%, of which 26% are glycine). Predicts the N-terminal signal peptide of GRP as an α-helix, while the rest of the sequence (tick-GRP77) remains unstructured, indicating a disordered region.
Figure \(\PageIndex{x}\) shows interactive iCn3D model of the AlphaFold structure of Tick saliva Glycine Rich Protein (Q4PME3). Glycines are shown in spacefill. Basic side chains (Arg, Lys) are shown in blue sticks, and the aromatic (Tyr and Phe) are shown in gray sticks.
Figure \(\PageIndex{x}\): AlphaFold structure of Tick saliva Glycine Rich Protein (Q4PME3). (Copyright; author via source). Click the image for a popup or use this external link: https://www.ncbi.nlm.nih.gov/Structu...656J4hCuHDDup9.
The first 19 amino acids, which form a predicted alpha-helix, form the signal peptide cleaved from the pro-protein to form the mature protein. The remaining portion of the protein (77 amino acids) is called tick-GRP77. The predicted AlphaFold model for GRP77 is completely disordered.
Tick saliva, on evaporation, becomes more concentrated and undergoes a liquid-liquid phase separation to form condensates. Results from lab studies of GRP77 are shown in Figure \(\PageIndex{x}\) below. Panel d shows the condensates. Panels a and b show key amino acid positions in the N-terminus (of the mature protein, the C-terminus, and mutants of the C-terminus in which the amino acids phenylalanine and tyrosine (ΔFY) or arginine (ΔR) have changed to alanine to probe the importance of F, Y, and R for aggregation.
Figure \(\PageIndex{x}\): Characteristics of peptides used in condensation studies. Ganar, K.A. et al. ibid
Panel d: Evaporation of tick-GRP77 on a surface-passivated glass slide showing numerous spherical tick-GRP77 condensates.
Panel a: The four fractions of tick-GRP77: N terminus (1–32), C terminus (33–77), C terminus mutant without aromatic amino acids (ΔFY mutant), and C terminus mutant without arginine residues (ΔR mutant). They all have a similar glycine content but differ in the number of basic (green), acidic (red), and aromatic (blue) amino acid residues. Panel b: Net charge per residue as a function of amino-acid position for the two termini, obtained using CIDER. The N terminus is strongly negatively charged, and the C terminus is moderately positively charged.
In vitro condensation experiments were conducted using droplet evaporation assays for the N-terminal and C-terminal peptides and respective mutants. It was found that the N-terminus formed underwent phase separation and condensation on a similar time scale as GRP77. The C-terminal separated quickly (9 min) when placed on specially treated surfaces. Since they differ in charge, it's unlikely that simple salt bridges mediate condensate formation. More likely, cation:pi (between Arg/Lys and the aromatic electron clouds of Tyr and Phe) and pi:pi interactions are key in aggregate formation and stabilization. Indeed, mutations that replaced aromatic side chains with the nonpolar amino acid alanine led to weak condensate formation. Likewise, replacing Arg with Ala reduced condensate formation.
Silent Single-nucleotide polymorphisms (SNPs)
For some amino acids, multiple triplet nucleotide sequences (codons) in the gene coding regions for a protein lead to the incorporation of the same amino acid in the protein sequence. Hence, two proteins identical in amino acid sequence might have slightly different nucleotide sequences in the encoded gene. Such single-nucleotide polymorphisms (SNPs) in coding regions were thought not to affect the tertiary structure and biological function of a protein if the single nucleotide variation did not lead to the insertion of a different amino acid into the growing peptide chain (i.e, the codons were synonymous and the mutations presumably silent with no effect). Recently, single-nucleotide polymorphisms (SNPs) in the MDR1 (multidrug resistance 1) gene, which encodes P-glycoprotein, were shown to result in a protein with altered substrate specificity and inhibitor interactions, and hence a different 3D structure. One possible explanation for this observation is a difference in the translation rate of the mRNA encoding this membrane protein. Different rates might yield different intra- and intermolecular associations, resulting in distinct final 3D structures as the protein cotranslationally folds and inserts into the membrane. This would be especially true if two possible structures were close enough in free energy but separated by a significant activation energy barrier, precluding simple conformational rearrangement between them.
It has been shown in yeast that synonymous mutations (those that don't change the amino acid on the mutation of the DNA encoding the particular amino acid) generally have the same effect on the "health" of yeast as do non-synonymous mutations (those that change the amino acid). This rather startling result upends much dogma. Some possible effects of synonymous mutation include alterations in gene expression of the mutated gene and effects on the stability of the transcribed RNA. Both types of mutations reduce mRNA levels and the fitness of the yeast, as defined by growth rate.
Metamorphic Proteins
In addition to prion proteins, many proteins can adopt multiple conformations under the same conditions. In contrast to prion proteins, for which the formation of the beta-structure variant is irreversible since the conformational change is associated with aggregation, many proteins can change conformations reversibly. Often, these changes are not associated solely with binding interactions that trigger them. Murzin has described proteins that change conformation upon changes in pH (viral glycoproteins), redox state (chloride channel), disulfide isomerization (lysozyme), and bound ligand (RNA polymerase as it initiates and then elongates the growing RNA polymer). He cites two proteins that appear to change state without external signals. These include Mad2, in which the two conformers share an extensive similarity, and Ltn10 (lymphotactin), in which they don't. One form of lymphotactin (Ltn 10) binds to similar lymphokine receptors, while the other (Ltn 40) binds to heparin. Folding kinetics may play a part in these examples as well, as proteins capable of folding to two conformers independently and quickly might prevent misfolding and aggregation that might occur if they had to unfold completely first before a conformational transition. Both Mad2 and Ltn10 alter conformation through transient dimer formation, facilitating conformational changes without widespread unfolding. Mutations in Ltn10 can cause the protein to adopt the Ltn40 conformation. Hence, primordial "metamorphic" proteins could, by simple mutation, produce new protein functionalities.
Metamorphic proteins, which undergo large structural changes, often involving shifts in hydrogen bonding and hence secondary structure, differ from simpler allosteric proteins with smaller conformational changes. Few metamorphic proteins have been found, but some speculate they could account for as much as 5% of proteins. An excellent example of such a protein is the human chemokine XCL1 (lymphotactin), an immune regulatory protein. It undergoes a major transition from a typical chemokine fold to a dimer with extensive beta structure. Lymphotactin lacks one of the two disulfide bonds found in other chemokines, allowing greater conformational flexibility.
Figure \(\PageIndex{11}\) shows interactive iCn3D models of the solution structures of monomeric XCL1 (lymphotactin), PDB 2HDM (left), and a dimeric form, PDB 2JP1 (right).
| Monomeric XCL1 (lymphotactin PDB 2HDM) | Dimeric XCL1 (lymphotactin PDB 2JP1) |
|
|
|
Summary
This chapter examines the dynamic world of intrinsically disordered proteins (IDPs) and metamorphic proteins, challenging the traditional view that proteins must adopt a single, well-defined structure to be functional. It begins by defining IDPs and intrinsically disordered regions (IDRs) as protein segments that, due to their enrichment in polar and charged residues and low hydrophobicity, fail to form a stable hydrophobic core. Despite this apparent lack of structure, many IDPs perform crucial regulatory roles, particularly in cell signaling and transcription, often through mechanisms such as coupled folding and binding. In this process, an otherwise unstructured protein adopts a more ordered conformation upon binding its partner, a concept that supports the idea of a dynamic “Protein Trinity,” in which ordered, collapsed, and extended states coexist and contribute to biological function.
The chapter further discusses the biological implications of disorder, highlighting how IDPs are predisposed to post-translational modifications and are subject to rapid degradation, which together help regulate their cellular concentrations and interactions. Detailed examples, including proteins like SIC1 and tau, illustrate how intrinsic disorder facilitates multiple binding interactions and functional versatility, while also predisposing some proteins to form pathological aggregates in diseases such as Alzheimer’s and Parkinson’s.
In addition to IDPs, the chapter introduces metamorphic proteins—those capable of adopting two or more distinct conformations under the same environmental conditions. Unlike prion proteins, the conformational transitions in metamorphic proteins are reversible and are not solely triggered by external ligands. Proteins such as lymphotactin and Mad2 serve as prime examples, demonstrating how alternative conformations can yield distinct functional outcomes.
Finally, the text highlights advances in experimental and computational techniques, including molecular dynamics simulations and AI-based prediction tools, which have expanded our understanding of the conformational ensembles of disordered proteins and the factors governing metamorphic transitions. This emerging perspective underscores the fluidity of protein structure. It challenges the conventional “one sequence – one structure – one function” paradigm, opening new avenues for drug targeting and functional regulation in complex biological systems.
References
f17f4df-605c-4388-88c2-25b0f000b0ed@2.
File:Chirality with hands.jpg. (2017, September 16). Wikimedia Commons, the free media repository. Retrieved 17:34, July 10, 2019 from commons.wikimedia.org/w/index.php?title=File:Chirality_with_hands.jpg&oldid=258750003.
Wikipedia contributors. (2019, July 6). Zwitterion. In Wikipedia, The Free Encyclopedia. Retrieved 21:48, July 10, 2019, from en.Wikipedia.org/w/index.php?title=Zwitterion&oldid=905089721
Wikipedia contributors. (2019, July 8). Absolute configuration. In Wikipedia, The Free Encyclopedia. Retrieved 15:28, July 14, 2019, from en.Wikipedia.org/w/index.php?title=Absolute_configuration&oldid=905412423
Structural Biochemistry/Enzyme/Active Site. (2019, July 1). Wikibooks, The Free Textbook Project. Retrieved 16:55, July 16, 2019 from en.wikibooks.org/w/index.php?title=Structural_Biochemistry/Enzyme/Active_Site&oldid=3555410.
Structural Biochemistry/Proteins. (2019, March 24). Wikibooks, The Free Textbook Project. Retrieved 19:16, July 18, 2019 from en.wikibooks.org/w/index.php?title=Structural_Biochemistry/Proteins&oldid=3529061.
Fujiwara, K., Toda, H., and Ikeguchi, M. (2012) Dependence of a α-helical and β-sheet amino acid propensities on teh overall protein fold type. BMC Structural Biology 12:18. Available at: https://bmcstructbiol.biomedcentral.com/track/pdf/10.1186/1472-6807-12-18
Wikipedia contributors. (2019, July 16). Keratin. In Wikipedia, The Free Encyclopedia. Retrieved 17:50, July 19, 2019, from en.Wikipedia.org/w/index.php?title=Keratin&oldid=906578340
Wikipedia contributors. (2019, July 13). Alpha-keratin. In Wikipedia, The Free Encyclopedia. Retrieved 18:17, July 19, 2019, from en.Wikipedia.org/w/index.php?title=Alpha-keratin&oldid=906117410
Open Learning Initiative. (2019) Integumentary Levels of Organization. Carnegie Mellon University. In Anatomy & Physiology. Available at: https://oli.cmu.edu/jcourse/webui/syllabus/module.do?context=4348901580020ca6010f804da8baf7ba.
Wikipedia contributors. (2019, July 16). Collagen. In Wikipedia, The Free Encyclopedia. Retrieved 03:42, July 20, 2019, from en.Wikipedia.org/w/index.php?title=Collagen&oldid=906509954
Wikipedia contributors. (2019, July 2). Rossmann fold. In Wikipedia, The Free Encyclopedia. Retrieved 16:01, July 20, 2019, from https://en.Wikipedia.org/w/index.php?title=Rossmann_fold&oldid=904468788
Wikipedia contributors. (2019, May 30). TIM barrel. In Wikipedia, The Free Encyclopedia. Retrieved 16:46, July 20, 2019, from en.Wikipedia.org/w/index.php?title=TIM_barrel&oldid=899459569
Wikipedia contributors. (2019, July 16). Protein folding. In Wikipedia, The Free Encyclopedia. Retrieved 18:30, July 20, 2019, from https://en.Wikipedia.org/w/index.php?title=Protein_folding&oldid=906604145
Wikipedia contributors. (2019, June 11). Globular protein. In Wikipedia, The Free Encyclopedia. Retrieved 18:49, July 20, 2019, from en.Wikipedia.org/w/index.php?title=Globular_protein&oldid=901360467
Wikipedia contributors. (2019, July 11). Intrinsically disordered proteins. In Wikipedia, The Free Encyclopedia. Retrieved 19:52, July 20, 2019, from en.Wikipedia.org/w/index.php?title=Intrinsically_disordered_proteins&oldid=905782287





.png?revision=1&size=bestfit&width=356&height=269)
.png?revision=1&size=bestfit&width=307&height=245)
.png?revision=1&size=bestfit&width=1044&height=183)


.png?revision=1&size=bestfit&width=321&height=302)
.png?revision=1)
.png?revision=1)