9.13: Ribozymes
- Page ID
- 4895
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Until about 20 years ago, all known enzymes were proteins. But then it was discovered that some RNA molecules can act as enzymes; that is, catalyze covalent changes in the structure of substrates (most of which are also RNA molecules). Catalytic RNA molecules are called ribozymes. Most classes of RNA, including transfer RNA (tRNA), ribosomal RNA (rRNA), and messenger RNA (mRNA) are transcribed as precursors that are larger than the final product. These precursors often contain "head" (5') and "tail" (3') sequences and intron sequences that must be removed to make the final product. Some of the processing steps employ other RNA molecules (always associated with proteins).
Ribonuclease P
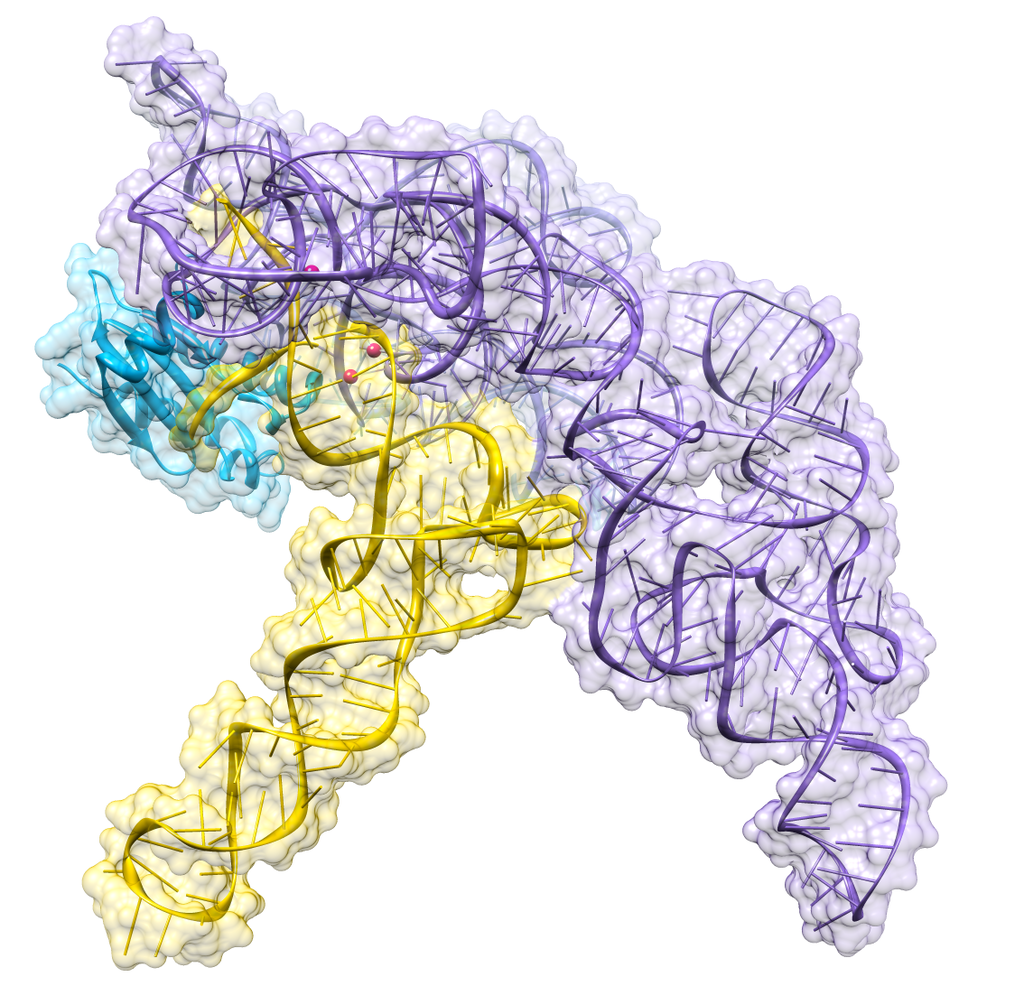
Almost all living things synthesize an enzyme — called Ribonuclease P (RNase P) — that cleaves the head (5') end of the precursors of transfer RNA (tRNA) molecules. In bacteria, ribonuclease P is a heterodimer containing a molecule of RNA and one of protein. Separated from each other, the RNA retains its ability to catalyze the cleavage step (although less efficiently than the intact dimer), but the protein alone cannot do the job.
Figure 9.13.1: Crystal structure of a bacterial ribonuclease P holoenzyme in complex with tRNA (yellow), showing metal ions involved in catalysis (pink spheres), PDB: 3Q1R. (CC BY SA 30; RNAMacGyver).
Group I Introns
Some ribosomal RNA (rRNA) genes, including those in the mitochondrial genome of certain fungi (e.g., yeast), in some chloroplast genomes and in the nuclear genome of some "lower" eukaryotes (e.g., the ciliated protozoan Tetrahymena thermophila and plasmodial slime moldPhysarum polycephalum) contain introns that must be spliced out to make the final product.
The splicing reaction is self-contained; that is, the intron — with the help of associated proteins — splices itself out of the precursor RNA. Once excision of the intron and splicing of the adjacent exons are completed, the story is over. In other words, although the action is catalyzed by the RNA, only a single molecule of substrate is involved (unlike protein enzymes that repeatedly catalyze a reaction).
However, synthetic versions of Group I introns made in the laboratory can — in vitro — act repeatedly; that is, like true enzymes. The DNA of some Group I introns includes an open reading frame (ORF) that encodes a transposase-like protein that can make a copy of the intron and insert it elsewhere in the genome. All the Group I introns share a characteristic secondary structure and mode of action that distinguishes them from the next group.
Group II Introns
Some messenger RNA (mRNA) genes in the mitochondrial genome of yeast and other fungi (encoding the proteins cytochrome b and subunits of cytochrome c oxidase) and in some chloroplast genomes also contain self-splicing introns. Because their secondary structure and the details of the splicing reaction differ from the rRNA introns discussed above, these are called Group II introns. The DNA of some Group II introns also includes an open reading frame (ORF) that encodes a transposase-like protein that can make a copy of the intron and insert it elsewhere in the genome.
Spliceosomes
Spliceosomes remove introns and splice the exons of most nuclear genes. They are composed of 5 kinds of small nuclear RNA (snRNA) molecules and over 100 different protein molecules. It is the RNA — not the protein — that catalyzes the splicing reactions. The molecular details of the reactions are similar to those of Group II introns, and this has led to speculation that this splicing machinery evolved from them.
Viroids
Viroids are DNA molecules that infect plant cells as conventional viruses do, but are far smaller (one has only 246 nucleotides). They are naked; that is, they are not encased in a capsi like viruses. Some viroidlike molecules get into the cell as passengers inside a conventional plant virus. These are called virusoids or viroidlike satellite RNAs.
In both cases, the molecules consists of single-stranded RNA whose ends are covalently bonded to form a circle. There are several regions where base-pairing occurs across adjacent portions of the molecule. New viroids and virusoids are synthesized by the host cell as long precursors in which the viroid structure is tandemly repeated. These repeats must be cut out and ligated to form the final product. Most virusoids and at least one viroid are self-splicing; that is, they can cut themselves out of the precursor and ligate their ends without the aid of any host enzymes. Thus they represent another class of ribozyme.
Both viroids and virusoids are responsible for a number of serious diseases of economically important plants, e.g. the coconut palm and chrysanthemums. (The problem is so severe with chrysanthemums that all growers in the U.S. now secure their stock from a few companies that raise the plants in "clean" rooms using stringent precautions to prevent infection by the viroid.)