3.4: Introducting Recombinant DNA into Host Cells
- Page ID
- 10524
Transformation in E. coli
E. coli does not have a natural system for taking up DNA, but when treated with \(\ce{CaCl2}\), the cells will take up the added DNA (Figure \(\PageIndex{1}\)). The recombinant vectors will give a new phenotype to the cells (usually drug resistance), so this process can be considered DNA-mediated transformation. An average efficiency is about 106 transformants per mg of DNA, although some more elaborate transformation cocktails procedures can give up to about 108 transformants per mg of DNA.
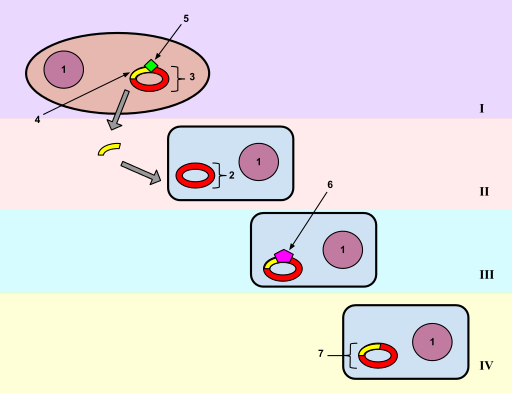
Usually one will transform with a mixture of recombinant vector molecules, most of which carry a different restriction fragment. Each transformed E. coli cell will pick up only one plasmid molecule, so the complex mixture of plasmids in the ligation mix has been separated into a population of transformed bacteria (Figre \(\PageIndex{1}\)). The bacterial cells are then plated at a sufficiently low density that individual colonies can be identified. Each colony (or transformant) carries a single plasmid, so as one screens the colonies, one is actually screening through individual DNA molecules. A colony is a visible group of bacterial cells on a plate, all of which are derived from a single bacterial cell. A group of identical cells derived from a single cell is called a clone. Since each clone carries a single type of recombinant DNA molecule, the process is called molecular cloning.
Phage Vectors
Phase vectors are a more efficient introduction of DNA into bacteria. Phage vectors such as those derived from bacteriophage l can carry larger inserts and can be introduced into bacteria more efficiently. l phage has a duplex DNA genome of about 50 kb. The internal 20 kb can be replaced with foreign DNA and still retain the lytic functions. Hence restriction fragments up to 20 kb can replace the l sequences, allowing larger genomic DNA fragments to be cloned (Figure \(\PageIndex{2}\)).
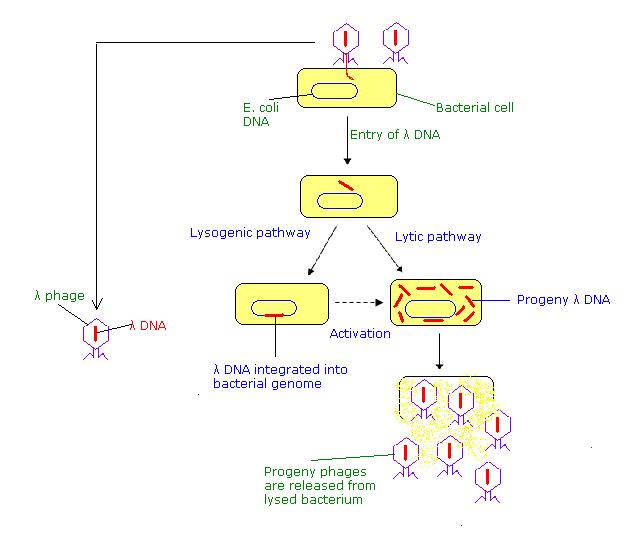
Recombinant bacteriophage can be introduction into E. coli by infection. DNA that has the cohesive ends of l can be packaged in vitro into infective phage particles. Being in a viral particle brings the efficiency of infection reliably over 108 plaque forming units per mg of recombinant DNA.
Some other bacteriphage vectors for cloning are derived from the virus M13. One can obtain single stranded DNA from M13 vectors and recombinants. M13 is a virus with a genome of single stranded DNA. It has a nonessential region into which foreign genes can be inserted. It has been modified to carry a gene for b‑galactosidase as a way to screen for recombinants. Introduction of recombinant M13 DNA into E. coli will lead to an infection of the host, and the progeny viral particles will contain single‑stranded DNA. The replicative form is duplex, allowing one to cleave with restriction enzymes and insert foreign DNA.
Some vectors are hybrids between plasmids and single‑strand phage; these are called phagemids. One example is pBluescript. Phagemids are plasmids (with the modified, high-copy number ColE1 origin) that also have an M13 origin of replication. Infection of transformed bacteria (containing the phagemid) with a helper virus (e.g. derived from M13) will cause the M13 origin to be activated, and progeny viruses carrying single‑stranded copies of the phagemid can be obtained. Hence one can easily obtain either double‑ or single‑stranded forms of thes plasmids. {The "blue" comes from the blue‑white screening for recombinants that can be done when the multiple cloning sites are in the b‑galactosidase gene. The "script" refers to the ability to make RNA copies of either strand in vitro with phage RNA polymerases.}
Vectors Designed to Carry Larger Inserts
Fragments even larger than those carried in l vectors are useful for studies of longer segments of chromosomes or whole genomes. Several vectors have been designed for cloning these very large fragments, 50 to 400 kb.
- Cosmids are plasmids that have the cohesive ends of l phage. They can be packaged in vitro into infective phage particles to give a more efficient delivery of the DNA into the cells. They can carry about 35 to 45 kb inserts.
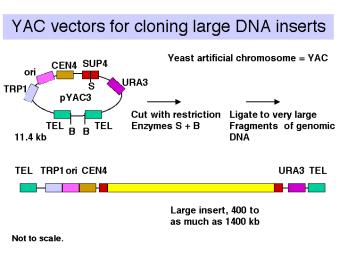
- Yeast artificial chromosomes (YACs) are yeast vectors with centromeres and telomeres. They can carry about 200 kb or larger fragments (in principle up to 1000 kb = 1 Mb). Thus very large fragments of DNA can be cloned in yeast (Figure \(\PageIndex{3}\)). In practice, chimeric clones with fragments from different regions of the genome are obtained fairly often, and some of the inserts are unstable.
 ,
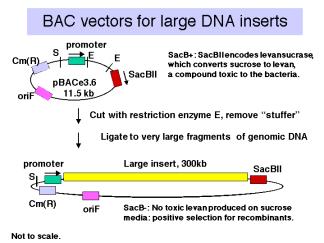
,Vectors derived from bacteriophage P1 can carry fragments of about 100 kb. Fragments in a similar size range are also cloned into bacterial artificial chromosomes (BACs), which are derived from the F-factor (Figure \(\PageIndex{4}\)). These have a lower copy number (like F) but they are stable and relatively easy to work with in the laboratory. BACs have become one of the most frequently used vectors for large inserts in genome projects.
Shuttle vectors for testing functions of isolated genes
Shuttle vectors can replicate in two different organisms, e.g. bacteria and yeast, or mammalian cells and bacteria. They have the appropriate origins of replication. Hence one can, e.g. clone a gene in bacteria, maybe modify it or mutate it in bacteria, and test its function by introducing it into yeast or animal cells.

