3.5: Polymerase Chain Reaction (PCR)
- Page ID
- 10525
The polymerase chain reaction (PCR) is now one of the most commonly used assays for obtaining a particular segment of DNA or RNA. It is rapid and extremely sensitive. By amplifying a designated segment of DNA, it provides a means to isolate that particular DNA segment or gene. This method requires knowledge of the nucleotide sequence at the ends of the region that you wish to amplify. Once that is known, one can make large quantities of that region starting with miniscule amounts of material, such as the DNA within a single human hair. With the availability of almost complete or complete sequences of genomes from many species, the range of genes to which it can be applied is enormous. The applications of PCR are numerous, from diagnostics to forensics to isolation of genes to studies of their expression.
The power of PCR lies in the exponential increase in amount of DNA that results from repeated cycles of DNA synthesis from primers that flank a given region, one primer designed to direct synthesis complementary to the top strand, the other designed to direct synthesis complementary to the bottom strand (Figure \(\PageIndex{1}\)). When this is done repeatedly, there is roughly a 2-fold increase in the amount of synthesized DNA in each cycle. Thus it is possible to generate a million-fold increase in the amount of DNA from the amplified region with a sufficient number of cycles. This exponential increase in abundance is similar to a chemical chain reaction, hence it is called the polymerase chain reaction.
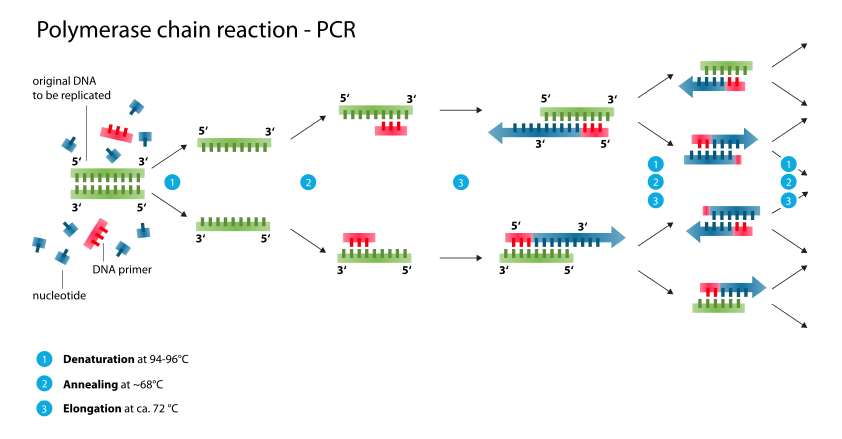
The events in the polymerase chain reaction are examined in more detail in Figure \(\PageIndex{2}\). The several panels show what happens in each cycle. Each cycle consists of a denaturation step at a temperature higher than the melting temperature of the duplex DNA (e.g. 95 oC ), then an annealing step at a temperature below the melting temperature for the primer-template (e.g. 55 oC), followed by extension of the primer by DNA polymerase using dNTPs provided in the reaction. This is done at the temperature optimum for the DNA polymerase (e.g. 70 oC for a thermostable polymerase). Thermocylers are commercially available for carrying out many cycles quickly and reliably (Figure \(\PageIndex{3}\)).
![image039[1].jpg](https://bio.libretexts.org/@api/deki/files/12059/image039%255B1%255D.jpg?revision=1)
![image037[1].jpg](https://bio.libretexts.org/@api/deki/files/12058/image037%255B1%255D.jpg?revision=1)
![image041[1].jpg](https://bio.libretexts.org/@api/deki/files/12060/image041%255B1%255D.jpg?revision=1)
![image043[1].jpg](https://bio.libretexts.org/@api/deki/files/12061/image043%255B1%255D.jpg?revision=1)
![image045[1].jpg](https://bio.libretexts.org/@api/deki/files/12062/image045%255B1%255D.jpg?revision=1)
The template supplied for the reaction is the only one available in the first cycle, and it is still a major template in the second cycle. At the end of the second cycle, a product is made whose ends are defined by primers. This is the desired product, and it serves as the major template for the remaining cycles. The initial template is still present and can be used, but it does not undergo the exponential expansion observed for the desire product.
If nis the number of cycles, the amount of desired product is approximately 2n-1 –2 times the amount of input DNA (between the primers). Thus in 21 cycles, one can achieve a million-fold increase in the amount of that DNA (assuming all cycles are completely efficient). A sample with 0.1 pg of the segment of DNA between the primers can be amplified to 0.1 mg in 21 cycles, in theory. In practice, roughly 25-35 cycles are done in many PCR assays.
The ease if doing PCR was greatly increased by the discovery of DNA polymerases that were stable at high temperatures. These have been isolated from bacteria that grow in hot springs, such as those found in Yellowstone National Park, such as Thermus aquaticus. The Taq polymerase from this bacterium will retain activity even at the high temperatures needed for melting the templates, and it is active at a temperature between the melting and annealing temperature. This particular polymerase is rather error-prone, and other thermostable polymerases have been discovered that are more accurate.


