mRNA Splicing
- Page ID
- 23915
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)1. Description of mRNA Splicing
Transcription and processing (which includes splicing) of the newly made mRNA occurs in the nucleus of the cell.
Once a mature mRNA transcript is made it is transported to the cytoplasm for translation into protein.
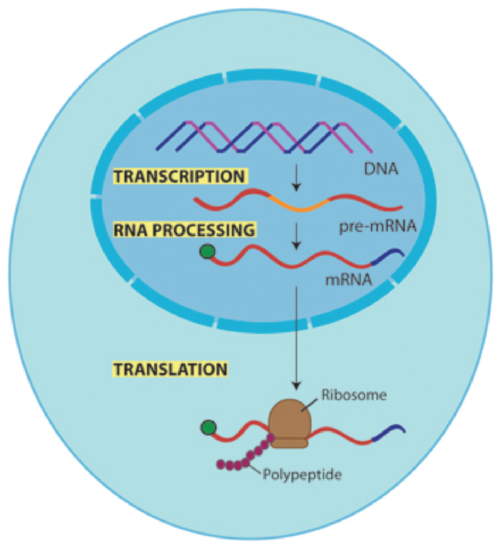
Figure \(\PageIndex{1}\). (CC BY-NC-SA)
Most eukaryotic genes and their pre-mRNA transcripts contain noncoding stretches of nucleotides or regions that are not meant to be made into protein. These noncoding segments are called intronsand must be removed before the mature mRNA can be transported to the cytoplasm and translated into protein. The stretches of DNA that do code for amino acids in the protein are called exons. During the process of splicing, introns are removed from the pre-mRNA by the spliceosome and exons are spliced back together. If the introns are not removed, the RNA would be translated into a nonfunctional protein. Splicing occurs in the nucleus before the RNA migrates to the cytoplasm. Once splicing is complete, the mature mRNA (containing uninterrupted coding information), is transported to the cytoplasm where ribosomes translate the mRNA into protein.
A Detailed Look at mRNA Splicing
The pre-mRNA Transcript
The pre-mRNA transcript contains both introns and exons. The introns are removed during the process of splicing. In this example, the pre-mRNA contains two exons and one intron.

Figure \(\PageIndex{2}\). (CC BY-NC-SA; Agathman)
Introns contain several important and conserved sequences that guide the splicing process: a 5’ GU sequence (the 5’ splice site), an A branch site located near a pyrimidine-rich region (a region with many cytosine and uracil bases) and a 3’ AG sequence (the 3’ splice site).

Figure \(\PageIndex{3}\). (CC BY-NC-SA; Agathman)
The Spliceosome
A large protein complex known as the spliceosome controls mRNA splicing. The spliceosome is composed of particles made up of both RNA and protein. These particles are called small nuclear ribonucleoprotein or snRNPs (pronounced “snurps”) for short. The snRNPs recognize the conserved sequences within introns and quickly bind these sequences once the pre-mRNA is made and initiate splicing.
The spliceosome is built in distinct steps. First, the U1 snRNP binds the 5’ splice site and the U2 snRNP binds the branch site.

Figure \(\PageIndex{4}\). (CC BY-NC-SA; Agathman)
A number of other snRNPs (U4, U6 and U5) bind the pre-mRNA transcript forming the mature spliceosome complex. This causes the intron to form a loop and brings the 5’ splice site and 3’ splice site together.

Figure \(\PageIndex{5}\). (CC BY-NC-SA; Agathman)
Now that the spliceosome is assembled, splicing can begin. First the 5’ end of the intron is cut. The 5’ GU end of the intron is then connected to the A branch site, which creates a lariat structure.

Figure \(\PageIndex{6}\). (CC BY-NC-SA; Agathman)
At this stage the U1 and U4 snRNPs are released and the 3’ splice site is cleaved. Once the intron has been fully cleaved, the two exons are attached to each other. The intron in the form of a lariat is released along with U2, U5 and U6 snRNPs.

Figure \(\PageIndex{7}\). (CC BY-NC-SA; Agathman)
The intron will be degraded and the snRNPs are used again to splice other pre-mRNAs. The mature mRNA transcript is now ready to be exported to the cytoplasm for translation.
Alternative Splicing
The example of a gene with a single intron and two exons used above is a very simple model of RNA splicing. Many genes contain multiple exons as well as multiple introns. A process known asalternative splicing allows for different combinations of exons to be included in the final mature mRNA, making different versions of proteins (called isoforms) that are all encoded by the same gene. Alternative splicing of mRNA allows for many proteins to be made, with different functions, all produced from a single gene. One of the most dramatic examples of alternative splicing is the Dscam gene in Drosophila melanogaster (a fruit fly). This single gene contains 116 exons! Some exons are always included, others may or may not be included. Over 18,000 different proteins from this single gene have been found in Drosophila! Theoretically, this system is capable of producing 38,016 different proteins all from a single gene!
Below is an example of alternative splicing of a pre-mRNA transcript. In this case, there are two different, alternatively spliced mRNAs that can be made from this pre-mRNA. The two mature mRNAs can contain either the yellow or the green exon. This produces two distinct protein isoforms when the mRNAs are translated into protein.

mRNA Splicing Tutorial by Dr. Katherine Harris is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License.
This tutorial was funded by the Title V-STEM Grant #P031S090007.


