3.3: Unique Characteristics of Prokaryotic Cells
- Page ID
- 5285
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Explain the distinguishing characteristics of prokaryotic cells
- Describe common cell morphologies and cellular arrangements typical of prokaryotic cells and explain how cells maintain their morphology
- Describe internal and external structures of prokaryotic cells in terms of their physical structure, chemical structure, and function
- Compare the distinguishing characteristics of bacterial and archaeal cells
Cell theory states that the cell is the fundamental unit of life. However, cells vary significantly in size, shape, structure, and function. At the simplest level of construction, all cells possess a few fundamental components. These include cytoplasm (a gel-like substance composed of water and dissolved chemicals needed for growth), which is contained within a plasma membrane (also called a cell membrane or cytoplasmic membrane); one or more chromosomes, which contain the genetic blueprints of the cell; and ribosomes, organelles used for the production of proteins.
Beyond these basic components, cells can vary greatly between organisms, and even within the same multicellular organism. The two largest categories of cells—prokaryotic cells and eukaryotic cells—are defined by major differences in several cell structures. Prokaryotic cells lack a nucleus surrounded by a complex nuclear membrane and generally have a single, circular chromosome located in a nucleoid. Eukaryotic cells have a nucleus surrounded by a complex nuclear membrane that contains multiple, rod-shaped chromosomes.1
All plant cells and animal cells are eukaryotic. Some microorganisms are composed of prokaryotic cells, whereas others are composed of eukaryotic cells. Prokaryotic microorganisms are classified within the domains Archaea and Bacteria, whereas eukaryotic organisms are classified within the domain Eukarya.
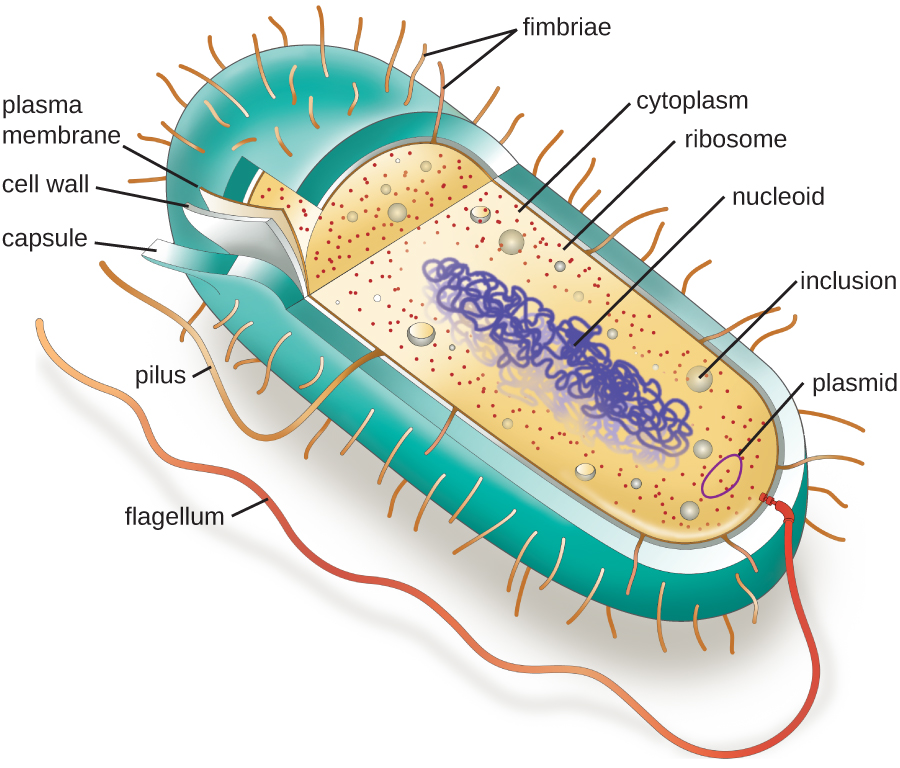
The structures inside a cell are analogous to the organs inside a human body, with unique structures suited to specific functions. Some of the structures found in prokaryotic cells are similar to those found in some eukaryotic cells; others are unique to prokaryotes. Although there are some exceptions, eukaryotic cells tend to be larger than prokaryotic cells. The comparatively larger size of eukaryotic cells dictates the need to compartmentalize various chemical processes within different areas of the cell, using complex membrane-bound organelles. In contrast, prokaryotic cells generally lack membrane-bound organelles; however, they often contain inclusions that compartmentalize their cytoplasm. Figure \(\PageIndex{1}\) illustrates structures typically associated with prokaryotic cells. These structures are described in more detail in the next section.
Common Cell Morphologies and Arrangements
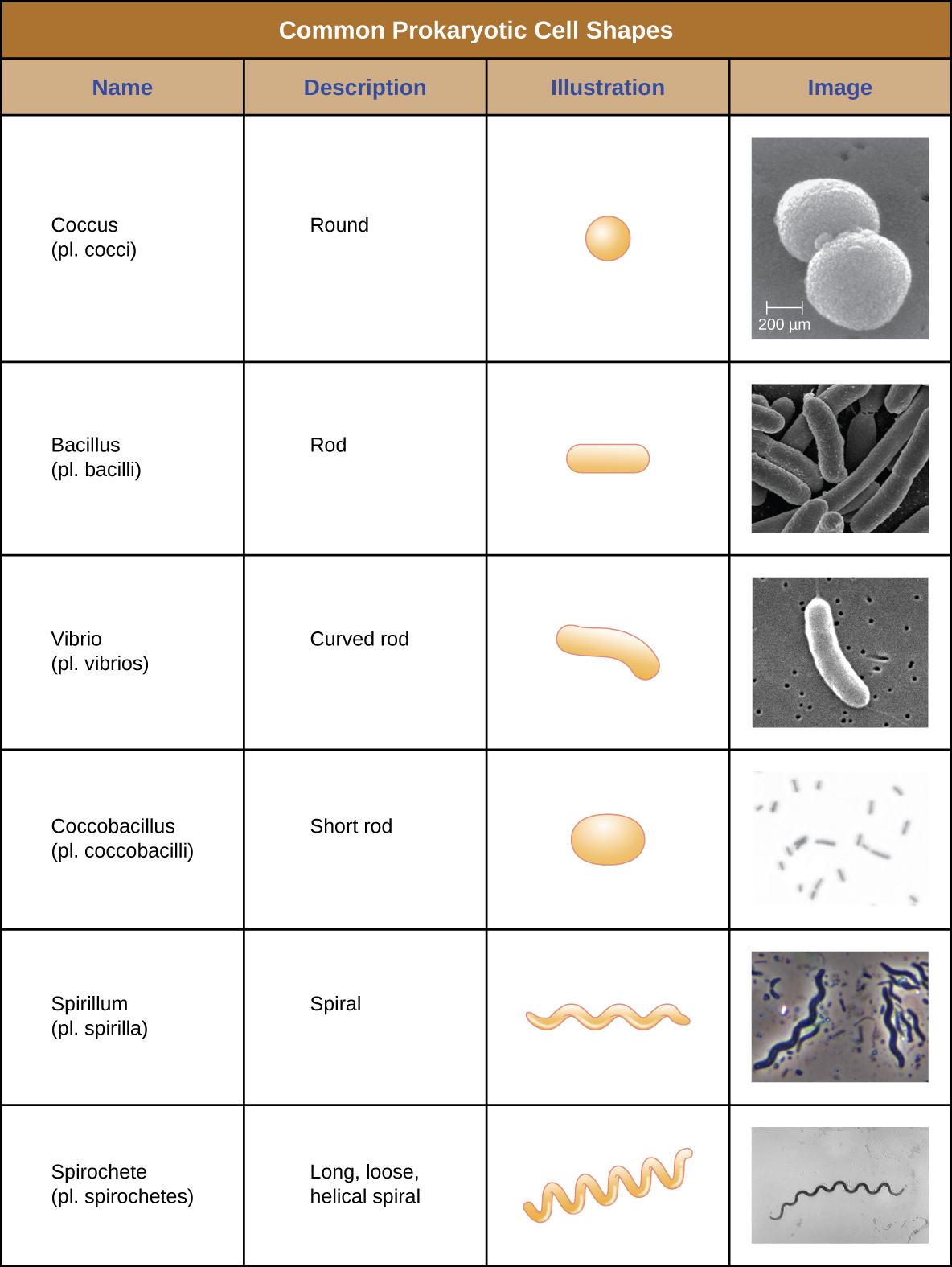
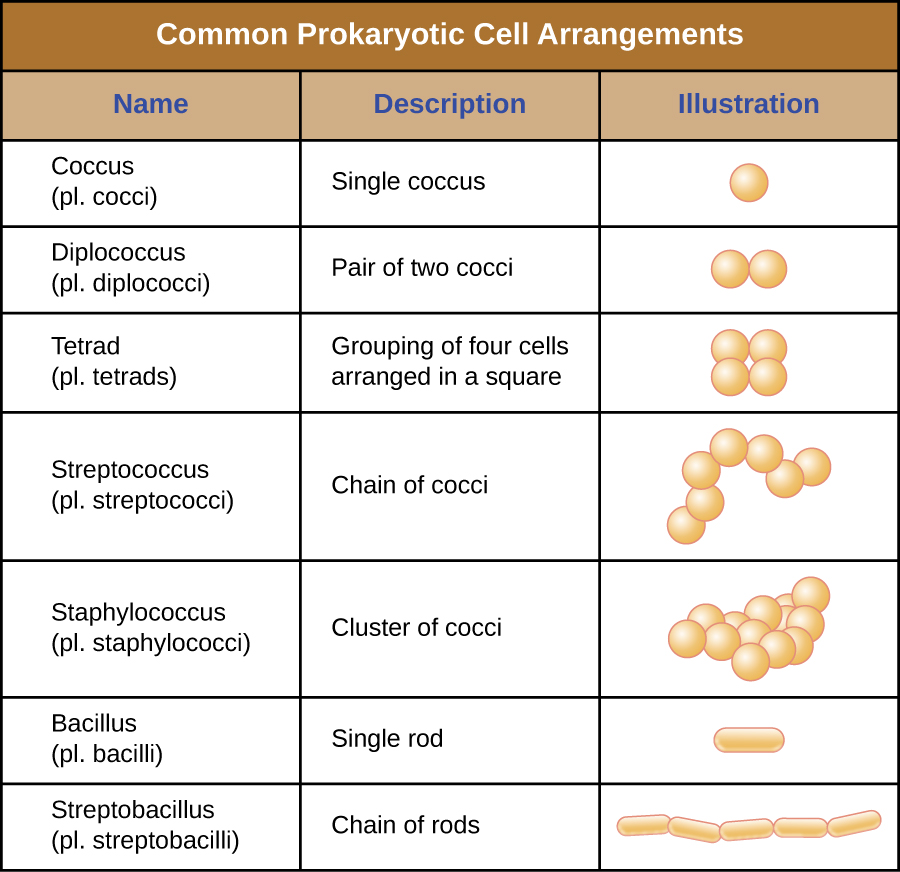
Individual cells of a particular prokaryotic organism are typically similar in shape, or cell morphology. Although thousands of prokaryotic organisms have been identified, only a handful of cell morphologies are commonly seen microscopically. Figure \(\PageIndex{2}\) names and illustrates cell morphologies commonly found in prokaryotic cells. In addition to cellular shape, prokaryotic cells of the same species may group together in certain distinctive arrangements depending on the plane of cell division. Some common arrangements are shown in Figure \(\PageIndex{3}\).
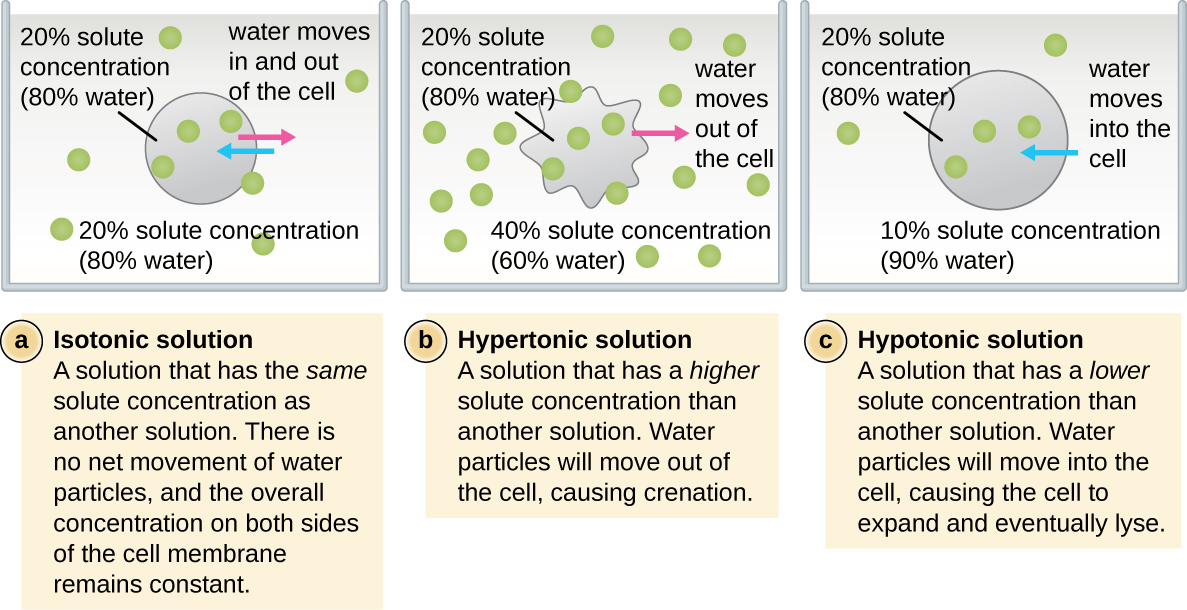
In most prokaryotic cells, morphology is maintained by the cell wall in combination with cytoskeletal elements. The cell wall is a structure found in most prokaryotes and some eukaryotes; it envelopes the cell membrane, protecting the cell from changes in osmotic pressure (Figure \(\PageIndex{4}\)). Osmotic pressure occurs because of differences in the concentration of solutes on opposing sides of a semipermeable membrane. Water is able to pass through a semipermeable membrane, but solutes (dissolved molecules like salts, sugars, and other compounds) cannot. When the concentration of solutes is greater on one side of the membrane, water diffuses across the membrane from the side with the lower concentration (more water) to the side with the higher concentration (less water) until the concentrations on both sides become equal. This diffusion of water is called osmosis, and it can cause extreme osmotic pressure on a cell when its external environment changes.
The external environment of a cell can be described as an isotonic, hypertonic, or hypotonic medium. In an isotonic medium, the solute concentrations inside and outside the cell are approximately equal, so there is no net movement of water across the cell membrane. In a hypertonic medium, the solute concentration outside the cell exceeds that inside the cell, so water diffuses out of the cell and into the external medium. In a hypotonic medium, the solute concentration inside the cell exceeds that outside of the cell, so water will move by osmosis into the cell. This causes the cell to swell and potentially lyse, or burst.
The degree to which a particular cell is able to withstand changes in osmotic pressure is called tonicity. Cells that have a cell wall are better able to withstand subtle changes in osmotic pressure and maintain their shape. In hypertonic environments, cells that lack a cell wall can become dehydrated, causing crenation, or shriveling of the cell; the plasma membrane contracts and appears scalloped or notched (Figure \(\PageIndex{4}\)). By contrast, cells that possess a cell wall undergo plasmolysis rather than crenation. In plasmolysis, the plasma membrane contracts and detaches from the cell wall, and there is a decrease in interior volume, but the cell wall remains intact, thus allowing the cell to maintain some shape and integrity for a period of time (Figure \(\PageIndex{5}\)). Likewise, cells that lack a cell wall are more prone to lysis in hypotonic environments. The presence of a cell wall allows the cell to maintain its shape and integrity for a longer time before lysing (Figure \(\PageIndex{5}\)).

- Explain the difference between cell morphology and arrangement.
- What advantages do cell walls provide prokaryotic cells?
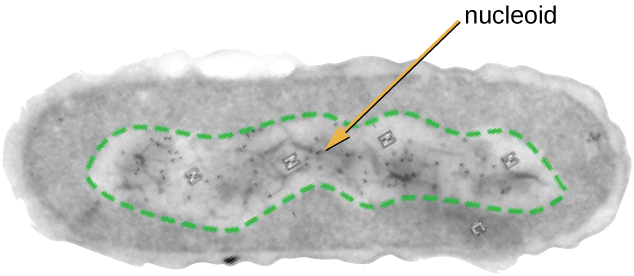
The Nucleoid
All cellular life has a DNA genome organized into one or more chromosomes. Prokaryotic chromosomes are typically circular, haploid (unpaired), and not bound by a complex nuclear membrane. Prokaryotic DNA and DNA-associated proteins are concentrated within the nucleoid region of the cell (Figure \(\PageIndex{6}\)). In general, prokaryotic DNA interacts with nucleoid-associated proteins (NAPs) that assist in the organization and packaging of the chromosome. In bacteria, NAPs function similar to histones, which are the DNA-organizing proteins found in eukaryotic cells. In archaea, the nucleoid is organized by either NAPs or histone-like DNA organizing proteins.
Plasmids
Prokaryotic cells may also contain extrachromosomal DNA, or DNA that is not part of the chromosome. This extrachromosomal DNA is found in plasmids, which are small, circular, double-stranded DNA molecules. Cells that have plasmids often have hundreds of them within a single cell. Plasmids are more commonly found in bacteria; however, plasmids have been found in archaea and eukaryotic organisms. Plasmids often carry genes that confer advantageous traits such as antibiotic resistance; thus, they are important to the survival of the organism. We will discuss plasmids in more detail in Mechanisms of Microbial Genetics.
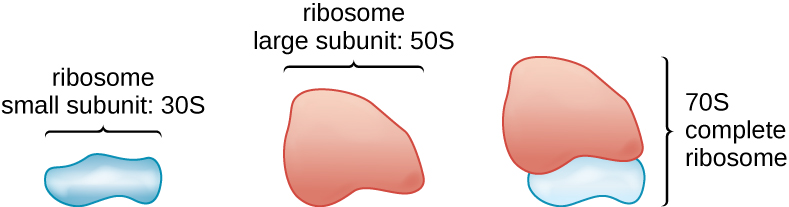
Ribosomes
All cellular life synthesizes proteins, and organisms in all three domains of life possess ribosomes, structures responsible protein synthesis. However, ribosomes in each of the three domains are structurally different. Ribosomes, themselves, are constructed from proteins, along with ribosomal RNA (rRNA). Prokaryotic ribosomes are found in the cytoplasm. They are called 70S ribosomes because they have a size of 70S (Figure \(\PageIndex{7}\)), whereas eukaryotic cytoplasmic ribosomes have a size of 80S. (The S stands for Svedberg unit, a measure of sedimentation in an ultracentrifuge, which is based on size, shape, and surface qualities of the structure being analyzed). Although they are the same size, bacterial and archaeal ribosomes have different proteins and rRNA molecules, and the archaeal versions are more similar to their eukaryotic counterparts than to those found in bacteria.
Inclusions
As single-celled organisms living in unstable environments, some prokaryotic cells have the ability to store excess nutrients within cytoplasmic structures called inclusions. Storing nutrients in a polymerized form is advantageous because it reduces the buildup of osmotic pressure that occurs as a cell accumulates solutes. Various types of inclusions store glycogen and starches, which contain carbon that cells can access for energy. Volutin granules, also called metachromatic granules because of their staining characteristics, are inclusions that store polymerized inorganic phosphate that can be used in metabolism and assist in the formation of biofilms. Microbes known to contain volutin granules include the archaea Methanosarcina, the bacterium Corynebacterium diphtheriae, and the unicellular eukaryotic alga Chlamydomonas. Sulfur granules, another type of inclusion, are found in sulfur bacteria of the genus Thiobacillus; these granules store elemental sulfur, which the bacteria use for metabolism.
Occasionally, certain types of inclusions are surrounded by a phospholipid monolayer embedded with protein. Polyhydroxybutyrate (PHB), which can be produced by species of Bacillus and Pseudomonas, is an example of an inclusion that displays this type of monolayer structure. Industrially, PHB has also been used as a source of biodegradable polymers for bioplastics. Several different types of inclusions are shown in Figure \(\PageIndex{8}\).
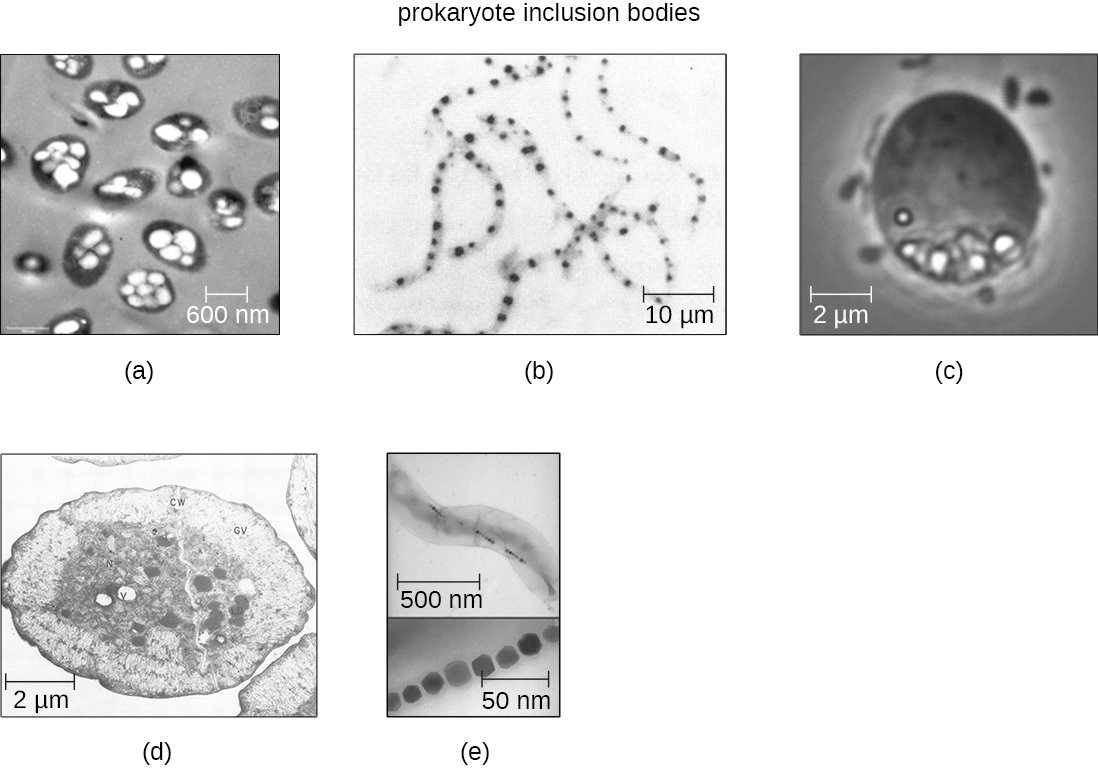
Some prokaryotic cells have other types of inclusions that serve purposes other than nutrient storage. For example, some prokaryotic cells produce gas vacuoles, accumulations of small, protein-lined vesicles of gas. These gas vacuoles allow the prokaryotic cells that synthesize them to alter their buoyancy so that they can adjust their location in the water column. Magnetotactic bacteria, such as Magnetospirillum magnetotacticum, contain magnetosomes, which are inclusions of magnetic iron oxide or iron sulfide surrounded by a lipid layer. These allow cells to align along a magnetic field, aiding their movement (Figure \(\PageIndex{8}\)). Cyanobacteria such as Anabaena cylindrica and bacteria such as Halothiobacillus neapolitanus produce carboxysome inclusions. Carboxysomes are composed of outer shells of thousands of protein subunits. Their interior is filled with ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) and carbonic anhydrase. Both of these compounds are used for carbon metabolism. Some prokaryotic cells also possess carboxysomes that sequester functionally related enzymes in one location. These structures are considered proto-organelles because they compartmentalize important compounds or chemical reactions, much like many eukaryotic organelles.
Endospores
Bacterial cells are generally observed as vegetative cells, but some genera of bacteria have the ability to form endospores, structures that essentially protect the bacterial genome in a dormant state when environmental conditions are unfavorable. Endospores (not to be confused with the reproductive spores formed by fungi) allow some bacterial cells to survive long periods without food or water, as well as exposure to chemicals, extreme temperatures, and even radiation. Table \(\PageIndex{1}\) compares the characteristics of vegetative cells and endospores.
| Vegetative Cells | Endospores |
|---|---|
| Sensitive to extreme temperatures and radiation | Resistant to extreme temperatures and radiation |
| Gram-positive | Do not absorb Gram stain, only special endospore stains (see Staining Microscopic Specimens) |
| Normal water content and enzymatic activity | Dehydrated; no metabolic activity |
| Capable of active growth and metabolism | Dormant; no growth or metabolic activity |
The process by which vegetative cells transform into endospores is called sporulation, and it generally begins when nutrients become depleted or environmental conditions become otherwise unfavorable (Figure \(\PageIndex{9}\)). The process begins with the formation of a septum in the vegetative bacterial cell. The septum divides the cell asymmetrically, separating a DNA forespore from the mother cell. The forespore, which will form the core of the endospore, is essentially a copy of the cell’s chromosomes, and is separated from the mother cell by a second membrane. A cortex gradually forms around the forespore by laying down layers of calcium and dipicolinic acid between membranes. A protein spore coat then forms around the cortex while the DNA of the mother cell disintegrates. Further maturation of the endospore occurs with the formation of an outermost exosporium. The endospore is released upon disintegration of the mother cell, completing sporulation.
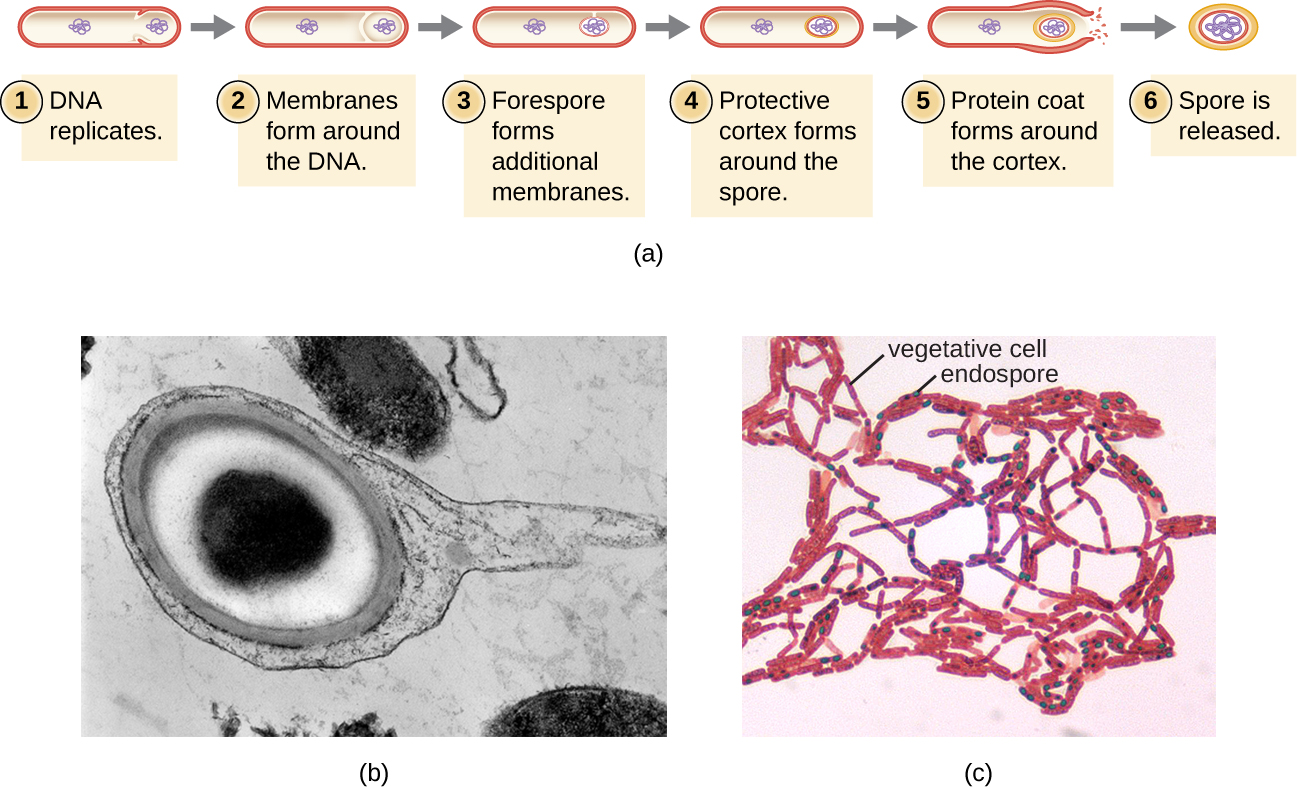
Endospores of certain species have been shown to persist in a dormant state for extended periods of time, up to thousands of years.2 However, when living conditions improve, endospores undergo germination, reentering a vegetative state. After germination, the cell becomes metabolically active again and is able to carry out all of its normal functions, including growth and cell division.
Not all bacteria have the ability to form endospores; however, there are a number of clinically significant endospore-forming gram-positive bacteria of the genera Bacillus and Clostridium. These include B. anthracis, the causative agent of anthrax, which produces endospores capable of survive for many decades3; C. tetani (causes tetanus); C. difficile (causes pseudomembranous colitis); C. perfringens (causes gas gangrene); and C. botulinum (causes botulism). Pathogens such as these are particularly difficult to combat because their endospores are so hard to kill. Special sterilization methods for endospore-forming bacteria are discussed in Control of Microbial Growth.
- What is an inclusion?
- What is the function of an endospore?
Plasma Membrane
Structures that enclose the cytoplasm and internal structures of the cell are known collectively as the cell envelope. In prokaryotic cells, the structures of the cell envelope vary depending on the type of cell and organism. Most (but not all) prokaryotic cells have a cell wall, but the makeup of this cell wall varies. All cells (prokaryotic and eukaryotic) have a plasma membrane (also called cytoplasmic membrane or cell membrane) that exhibits selective permeability, allowing some molecules to enter or leave the cell while restricting the passage of others.
The structure of the plasma membrane is often described in terms of the fluid mosaic model, which refers to the ability of membrane components to move fluidly within the plane of the membrane, as well as the mosaic-like composition of the components, which include a diverse array of lipid and protein components (Figure \(\PageIndex{10}\)). The plasma membrane structure of most bacterial and eukaryotic cell types is a bilayer composed mainly of phospholipids formed with ester linkages and proteins. These phospholipids and proteins have the ability to move laterally within the plane of the membranes as well as between the two phospholipid layers.
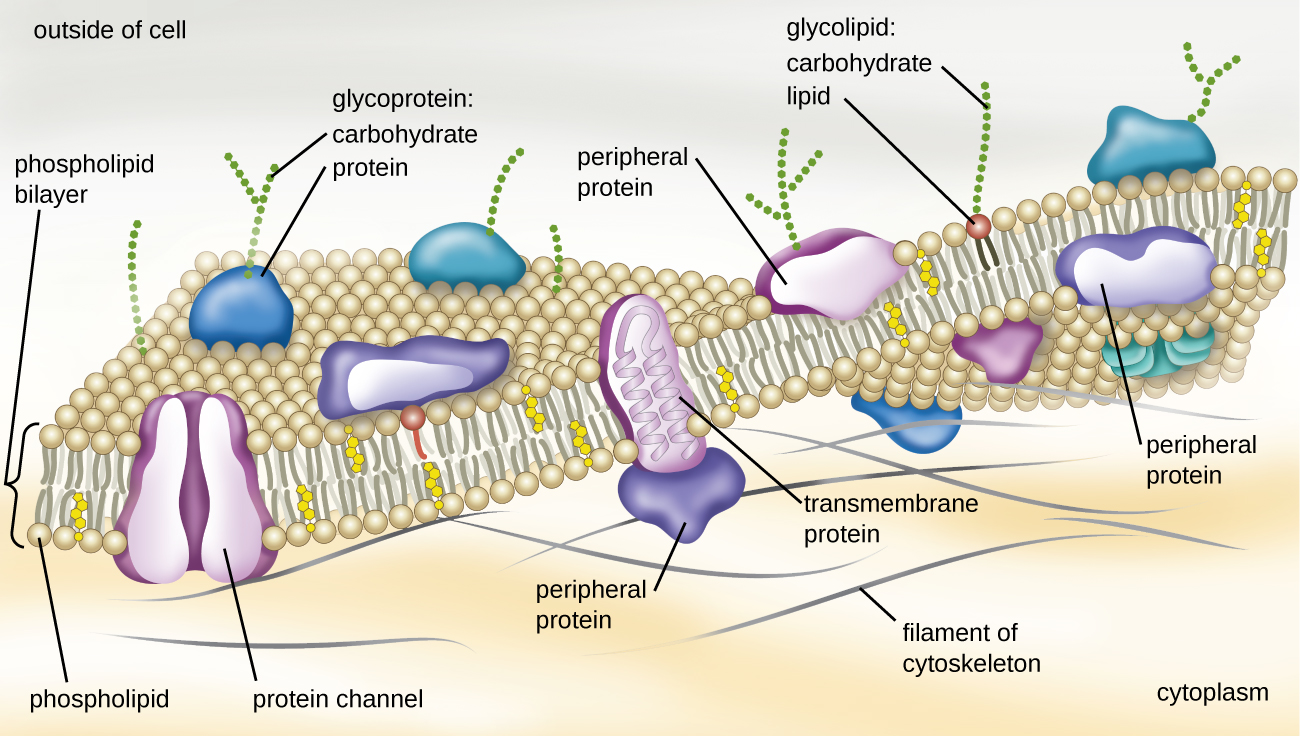
Archaeal membranes are fundamentally different from bacterial and eukaryotic membranes in a few significant ways. First, archaeal membrane phospholipids are formed with ether linkages, in contrast to the ester linkages found in bacterial or eukaryotic cell membranes. Second, archaeal phospholipids have branched chains, whereas those of bacterial and eukaryotic cells are straight chained. Finally, although some archaeal membranes can be formed of bilayers like those found in bacteria and eukaryotes, other archaeal plasma membranes are lipid monolayers.
Proteins on the cell’s surface are important for a variety of functions, including cell-to-cell communication, and sensing environmental conditions and pathogenic virulence factors. Membrane proteins and phospholipids may have carbohydrates (sugars) associated with them and are called glycoproteins or glycolipids, respectively. These glycoprotein and glycolipid complexes extend out from the surface of the cell, allowing the cell to interact with the external environment (Figure \(\PageIndex{10}\)). Glycoproteins and glycolipids in the plasma membrane can vary considerably in chemical composition among archaea, bacteria, and eukaryotes, allowing scientists to use them to characterize unique species.
Plasma membranes from different cells types also contain unique phospholipids, which contain fatty acids. As described in Using Biochemistry to Identify Microorganisms, phospholipid-derived fatty acid analysis (PLFA) profiles can be used to identify unique types of cells based on differences in fatty acids. Archaea, bacteria, and eukaryotes each have a unique PFLA profile.
Membrane Transport Mechanisms
One of the most important functions of the plasma membrane is to control the transport of molecules into and out of the cell. Internal conditions must be maintained within a certain range despite any changes in the external environment. The transport of substances across the plasma membrane allows cells to do so.
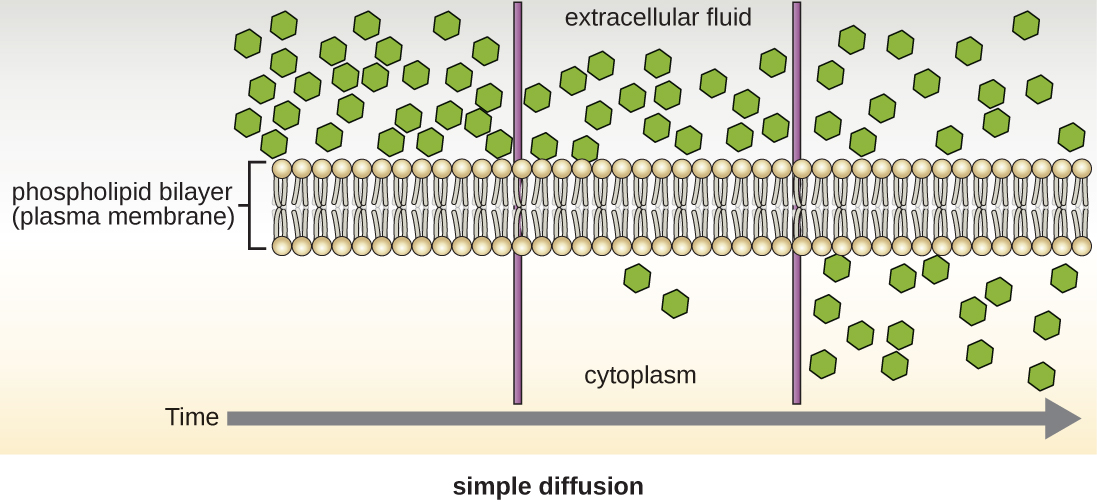
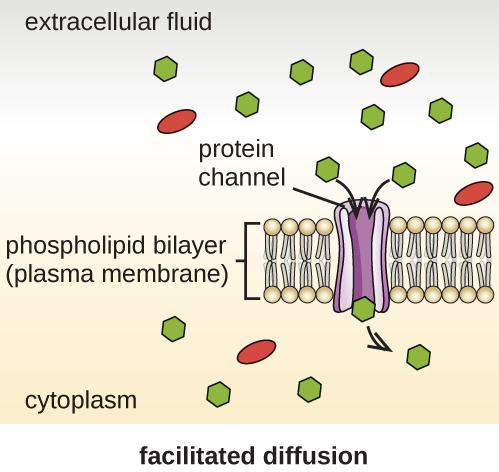
Cells use various modes of transport across the plasma membrane. For example, molecules moving from a higher concentration to a lower concentration with the concentration gradient are transported by simple diffusion, also known as passive transport (Figure \(\PageIndex{11}\)). Some small molecules, like carbon dioxide, may cross the membrane bilayer directly by simple diffusion. However, charged molecules, as well as large molecules, need the help of carriers or channels in the membrane. These structures ferry molecules across the membrane, a process known as facilitated diffusion (Figure \(\PageIndex{12}\)).
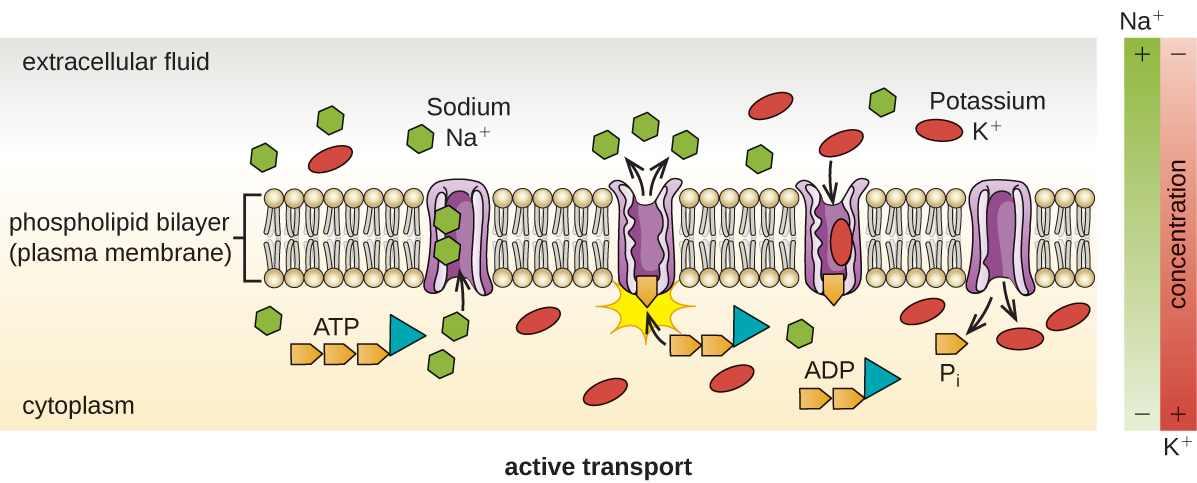
Active transport occurs when cells move molecules across their membrane against concentration gradients (Figure \(\PageIndex{13}\)). A major difference between passive and active transport is that active transport requires adenosine triphosphate (ATP) or other forms of energy to move molecules “uphill.” Therefore, active transport structures are often called “pumps.”
Group translocation also transports substances into bacterial cells. In this case, as a molecule moves into a cell against its concentration gradient, it is chemically modified so that it does not require transport against an unfavorable concentration gradient. A common example of this is the bacterial phosphotransferase system, a series of carriers that phosphorylates (i.e., adds phosphate ions to) glucose or other sugars upon entry into cells. Since the phosphorylation of sugars is required during the early stages of sugar metabolism, the phosphotransferase system is considered to be an energy neutral system.
Photosynthetic Membrane Structures
Some prokaryotic cells, namely cyanobacteria and photosynthetic bacteria, have membrane structures that enable them to perform photosynthesis. These structures consist of an infolding of the plasma membrane that encloses photosynthetic pigments such as green chlorophylls and bacteriochlorophylls. In cyanobacteria, these membrane structures are called thylakoids; in photosynthetic bacteria, they are called chromatophores, lamellae, or chlorosomes.
Cell Wall
The primary function of the cell wall is to protect the cell from harsh conditions in the outside environment. When present, there are notable similarities and differences among the cell walls of archaea, bacteria, and eukaryotes.
The major component of bacterial cell walls is called peptidoglycan (or murein); it is only found in bacteria. Structurally, peptidoglycan resembles a layer of meshwork or fabric (Figure \(\PageIndex{14}\)). Each layer is composed of long chains of alternating molecules of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). The structure of the long chains has significant two-dimensional tensile strength due to the formation of peptide bridges that connect NAG and NAM within each peptidoglycan layer. In gram-negative bacteria, tetrapeptide chains extending from each NAM unit are directly cross-linked, whereas in gram-positive bacteria, these tetrapeptide chains are linked by pentaglycine cross-bridges. Peptidoglycan subunits are made inside of the bacterial cell and then exported and assembled in layers, giving the cell its shape.
Since peptidoglycan is unique to bacteria, many antibiotic drugs are designed to interfere with peptidoglycan synthesis, weakening the cell wall and making bacterial cells more susceptible to the effects of osmotic pressure (see Mechanisms of Antibacterial Drugs). In addition, certain cells of the human immune system are able “recognize” bacterial pathogens by detecting peptidoglycan on the surface of a bacterial cell; these cells then engulf and destroy the bacterial cell, using enzymes such as lysozyme, which breaks down and digests the peptidoglycan in their cell walls (see Pathogen Recognition and Phagocytosis).
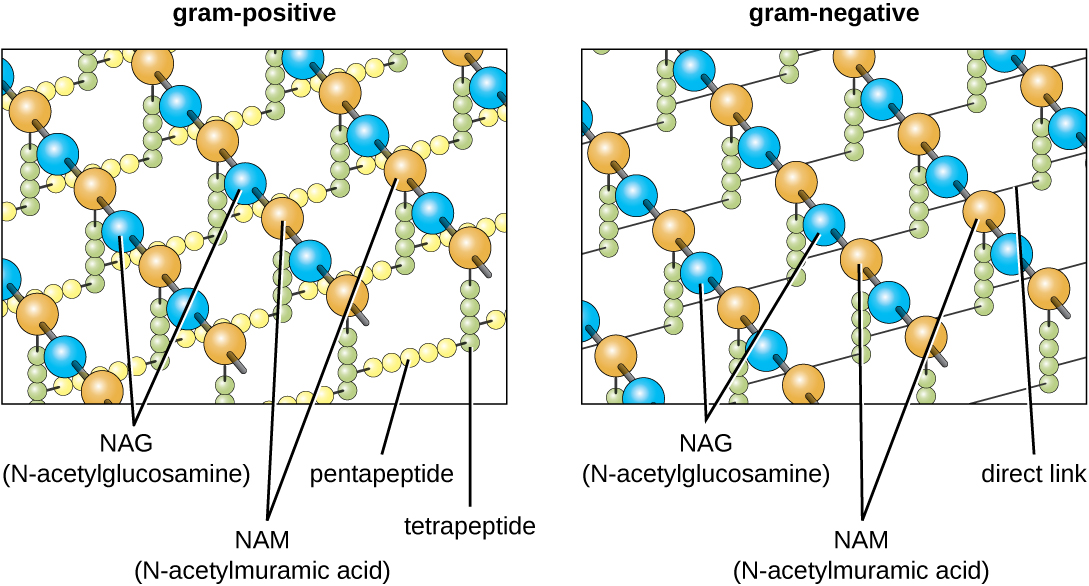
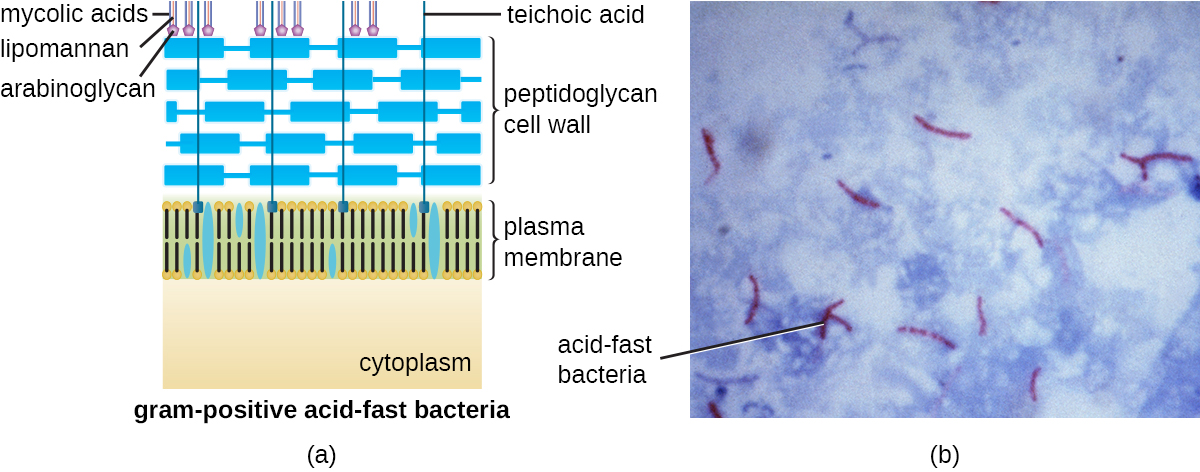
The Gram staining protocol (see Staining Microscopic Specimens) is used to differentiate two common types of cell wall structures (Figure \(\PageIndex{15}\)). Gram-positive cells have a cell wall consisting of many layers of peptidoglycan totaling 30–100 nm in thickness. These peptidoglycan layers are commonly embedded with teichoic acids (TAs), carbohydrate chains that extend through and beyond the peptidoglycan layer.4 TA is thought to stabilize peptidoglycan by increasing its rigidity. TA also plays a role in the ability of pathogenic gram-positive bacteria such as Streptococcus to bind to certain proteins on the surface of host cells, enhancing their ability to cause infection. In addition to peptidoglycan and TAs, bacteria of the family Mycobacteriaceae have an external layer of waxy mycolic acids in their cell wall; as described in Staining Microscopic Specimens, these bacteria are referred to as acid-fast, since acid-fast stains must be used to penetrate the mycolic acid layer for purposes of microscopy (Figure \(\PageIndex{16}\)).

Gram-negative cells have a much thinner layer of peptidoglycan (no more than about 4 nm thick6) than gram-positive cells, and the overall structure of their cell envelope is more complex. In gram-negative cells, a gel-like matrix occupies the periplasmic space between the cell wall and the plasma membrane, and there is a second lipid bilayer called the outer membrane, which is external to the peptidoglycan layer (Figure \(\PageIndex{15}\)). This outer membrane is attached to the peptidoglycan by murein lipoprotein. The outer leaflet of the outer membrane contains the molecule lipopolysaccharide (LPS), which functions as an endotoxin in infections involving gram-negative bacteria, contributing to symptoms such as fever, hemorrhaging, and septic shock. Each LPS molecule is composed of Lipid A, a core polysaccharide, and an O side chain that is composed of sugar-like molecules that comprise the external face of the LPS (Figure \(\PageIndex{17}\)). The composition of the O side chain varies between different species and strains of bacteria. Parts of the O side chain called antigens can be detected using serological or immunological tests to identify specific pathogenic strains like Escherichia coli O157:H7, a deadly strain of bacteria that causes bloody diarrhea and kidney failure.

Archaeal cell wall structure differs from that of bacteria in several significant ways. First, archaeal cell walls do not contain peptidoglycan; instead, they contain a similar polymer called pseudopeptidoglycan (pseudomurein) in which NAM is replaced with a different subunit. Other archaea may have a layer of glycoproteins or polysaccharides that serves as the cell wall instead of pseudopeptidoglycan. Last, as is the case with some bacterial species, there are a few archaea that appear to lack cell walls entirely.
Glycocalyces and S-Layers
Although most prokaryotic cells have cell walls, some may have additional cell envelope structures exterior to the cell wall, such as glycocalyces and S-layers. A glycocalyx is a sugar coat, of which there are two important types: capsules and slime layers. A capsule is an organized layer located outside of the cell wall and usually composed of polysaccharides or proteins (Figure \(\PageIndex{18}\)). A slime layer is a less tightly organized layer that is only loosely attached to the cell wall and can be more easily washed off. Slime layers may be composed of polysaccharides, glycoproteins, or glycolipids.
Glycocalyces allows cells to adhere to surfaces, aiding in the formation of biofilms (colonies of microbes that form in layers on surfaces). In nature, most microbes live in mixed communities within biofilms, partly because the biofilm affords them some level of protection. Biofilms generally hold water like a sponge, preventing desiccation. They also protect cells from predation and hinder the action of antibiotics and disinfectants. All of these properties are advantageous to the microbes living in a biofilm, but they present challenges in a clinical setting, where the goal is often to eliminate microbes.
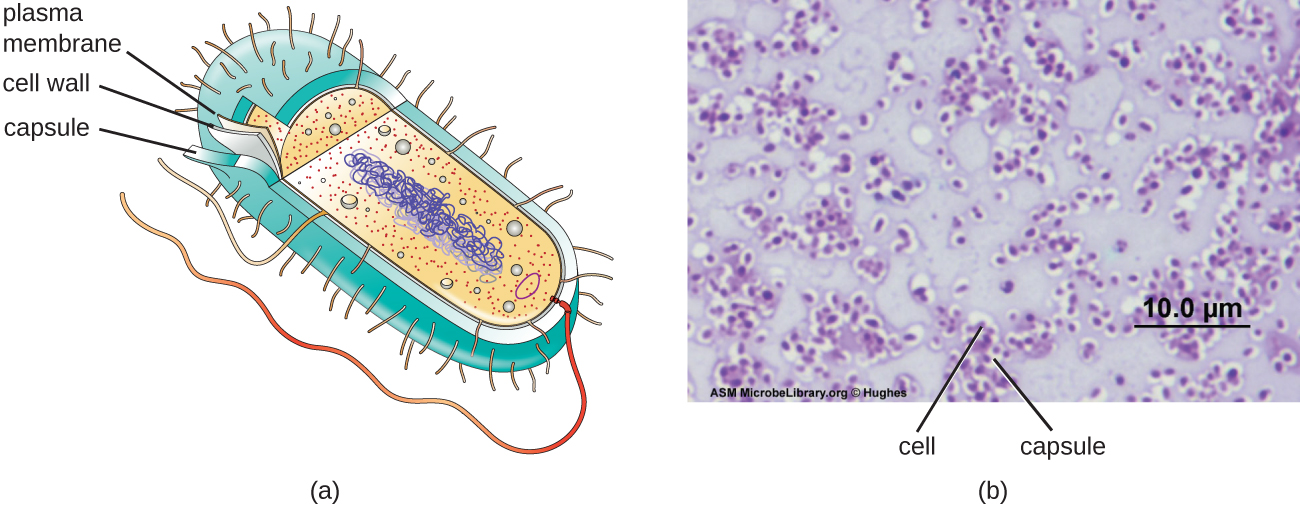
The ability to produce a capsule can contribute to a microbe’s pathogenicity (ability to cause disease) because the capsule can make it more difficult for phagocytic cells (such as white blood cells) to engulf and kill the microorganism. Streptococcus pneumoniae, for example, produces a capsule that is well known to aid in this bacterium’s pathogenicity. As explained in Staining Microscopic Specimens, capsules are difficult to stain for microscopy; negative staining techniques are typically used.
An S-layer is another type of cell envelope structure; it is composed of a mixture of structural proteins and glycoproteins. In bacteria, S-layers are found outside the cell wall, but in some archaea, the S-layer serves as the cell wall. The exact function of S-layers is not entirely understood, and they are difficult to study; but available evidence suggests that they may play a variety of functions in different prokaryotic cells, such as helping the cell withstand osmotic pressure and, for certain pathogens, interacting with the host immune system.
After diagnosing Barbara with pneumonia, the PA writes her a prescription for amoxicillin, a commonly-prescribed type of penicillin derivative. More than a week later, despite taking the full course as directed, Barbara still feels weak and is not fully recovered, although she is still able to get through her daily activities. She returns to the health center for a follow-up visit.
Many types of bacteria, fungi, and viruses can cause pneumonia. Amoxicillin targets the peptidoglycan of bacterial cell walls. Since the amoxicillin has not resolved Barbara’s symptoms, the PA concludes that the causative agent probably lacks peptidoglycan, meaning that the pathogen could be a virus, a fungus, or a bacterium that lacks peptidoglycan. Another possibility is that the pathogen is a bacterium containing peptidoglycan but has developed resistance to amoxicillin.
- How can the PA definitively identify the cause of Barbara’s pneumonia?
- What form of treatment should the PA prescribe, given that the amoxicillin was ineffective?
Filamentous Appendages
Many bacterial cells have protein appendages embedded within their cell envelopes that extend outward, allowing interaction with the environment. These appendages can attach to other surfaces, transfer DNA, or provide movement. Filamentous appendages include fimbriae, pili, and flagella.
Fimbriae and Pili
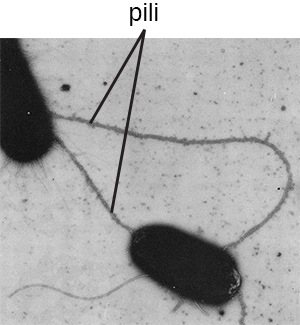
Fimbriae and pili are structurally similar and, because differentiation between the two is problematic, these terms are often used interchangeably.7 8 The term fimbriae commonly refers to short bristle-like proteins projecting from the cell surface by the hundreds. Fimbriae enable a cell to attach to surfaces and to other cells. For pathogenic bacteria, adherence to host cells is important for colonization, infectivity, and virulence. Adherence to surfaces is also important in biofilm formation.
The term pili (singular: pilus) commonly refers to longer, less numerous protein appendages that aid in attachment to surfaces (Figure \(\PageIndex{19}\)). A specific type of pilus, called the F pilus or sex pilus, is important in the transfer of DNA between bacterial cells, which occurs between members of the same generation when two cells physically transfer or exchange parts of their respective genomes (see How Asexual Prokaryotes Achieve Genetic Diversity).
Before the structure and function of the various components of the bacterial cell envelope were well understood, scientists were already using cell envelope characteristics to classify bacteria. In 1933, Rebecca Lancefield proposed a method for serotyping various β-hemolytic strains of Streptococcus species using an agglutination assay, a technique using the clumping of bacteria to detect specific cell-surface antigens. In doing so, Lancefield discovered that one group of S. pyogenes, found in Group A, was associated with a variety of human diseases. She determined that various strains of Group A strep could be distinguished from each other based on variations in specific cell surface proteins that she named M proteins.
Today, more than 80 different strains of Group A strep have been identified based on M proteins. Various strains of Group A strep are associated with a wide variety of human infections, including streptococcal pharyngitis (strep throat), impetigo, toxic shock syndrome, scarlet fever, rheumatic fever, and necrotizing fasciitis. The M protein is an important virulence factor for Group A strep, helping these strains evade the immune system. Changes in M proteins appear to alter the infectivity of a particular strain of Group A strep.
Flagella
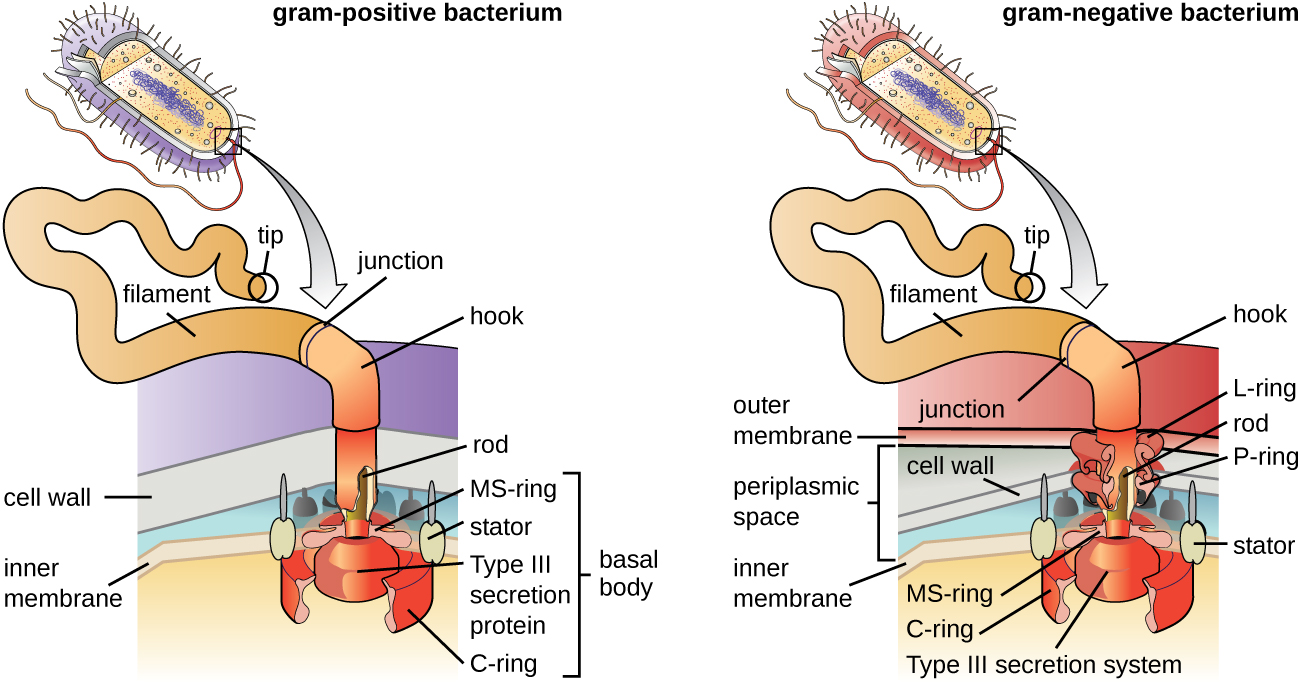
Flagella are structures used by cells to move in aqueous environments. Bacterial flagella act like propellers. They are stiff spiral filaments composed of flagellin protein subunits that extend outward from the cell and spin in solution. The basal body is the motor for the flagellum and is embedded in the plasma membrane (Figure \(\PageIndex{20}\)). A hook region connects the basal body to the filament. Gram-positive and gram-negative bacteria have different basal body configurations due to differences in cell wall structure.
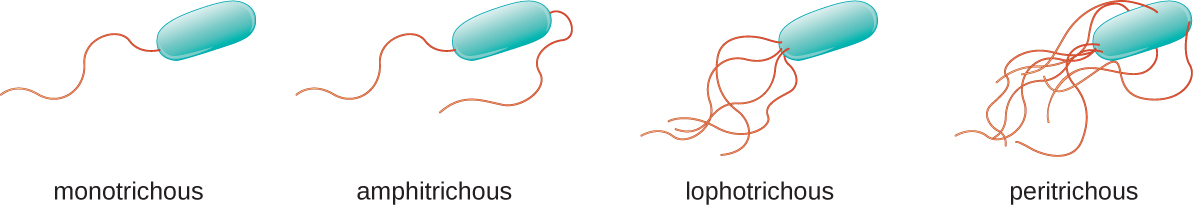
Different types of motile bacteria exhibit different arrangements of flagella (Figure \(\PageIndex{21}\)). A bacterium with a singular flagellum, typically located at one end of the cell (polar), is said to have a monotrichous flagellum. An example of a monotrichously flagellated bacterial pathogen is Vibrio cholerae, the gram-negative bacterium that causes cholera. Cells with amphitrichous flagella have a flagellum or tufts of flagella at each end. An example is Spirillum minor, the cause of spirillary (Asian) rat-bite fever or sodoku. Cells with lophotrichous flagella have a tuft at one end of the cell. The gram-negative bacillus Pseudomonas aeruginosa, an opportunistic pathogen known for causing many infections, including “swimmer’s ear” and burn wound infections, has lophotrichous flagella. Flagella that cover the entire surface of a bacterial cell are called peritrichous flagella. The gram-negative bacterium E. coli shows a peritrichous arrangement of flagella.
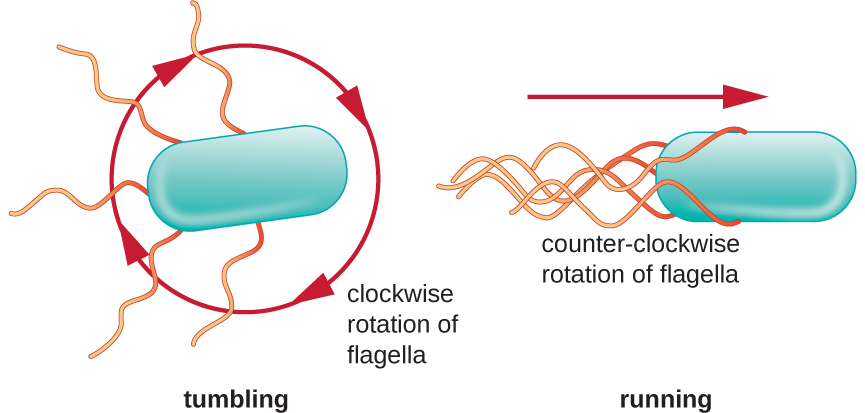
Directional movement depends on the configuration of the flagella. Bacteria can move in response to a variety of environmental signals, including light (phototaxis), magnetic fields (magnetotaxis) using magnetosomes, and, most commonly, chemical gradients (chemotaxis). Purposeful movement toward a chemical attractant, like a food source, or away from a repellent, like a poisonous chemical, is achieved by increasing the length of runs and decreasing the length of tumbles. When running, flagella rotate in a counterclockwise direction, allowing the bacterial cell to move forward. In a peritrichous bacterium, the flagella are all bundled together in a very streamlined way (Figure \(\PageIndex{22}\)), allowing for efficient movement. When tumbling, flagella are splayed out while rotating in a clockwise direction, creating a looping motion and preventing meaningful forward movement but reorienting the cell toward the direction of the attractant. When an attractant exists, runs and tumbles still occur; however, the length of runs is longer, while the length of the tumbles is reduced, allowing overall movement toward the higher concentration of the attractant. When no chemical gradient exists, the lengths of runs and tumbles are more equal, and overall movement is more random (Figure \(\PageIndex{23}\)).

- What is the peptidoglycan layer and how does it differ between gram-positive and gram-negative bacteria?
- Compare and contrast monotrichous, amphitrichous, lophotrichous, and peritrichous flagella.
Summary
- Prokaryotic cells differ from eukaryotic cells in that their genetic material is contained in a nucleoid rather than a membrane-bound nucleus. In addition, prokaryotic cells generally lack membrane-bound organelles.
- Prokaryotic cells of the same species typically share a similar cell morphology and cellular arrangement.
- Most prokaryotic cells have a cell wall that helps the organism maintain cellular morphology and protects it against changes in osmotic pressure.
- Outside of the nucleoid, prokaryotic cells may contain extrachromosomal DNA in plasmids.
- Prokaryotic ribosomes that are found in the cytoplasm have a size of 70S.
- Some prokaryotic cells have inclusions that store nutrients or chemicals for other uses.
- Some prokaryotic cells are able to form endospores through sporulation to survive in a dormant state when conditions are unfavorable. Endospores can germinate, transforming back into vegetative cells when conditions improve.
- In prokaryotic cells, the cell envelope includes a plasma membrane and usually a cell wall.
- Bacterial membranes are composed of phospholipids with integral or peripheral proteins. The fatty acid components of these phospholipids are ester-linked and are often used to identify specific types of bacteria. The proteins serve a variety of functions, including transport, cell-to-cell communication, and sensing environmental conditions. Archaeal membranes are distinct in that they are composed of fatty acids that are ether-linked to phospholipids.
- Some molecules can move across the bacterial membrane by simple diffusion, but most large molecules must be actively transported through membrane structures using cellular energy.
- Prokaryotic cell walls may be composed of peptidoglycan (bacteria) or pseudopeptidoglycan (archaea).
- Gram-positive bacterial cells are characterized by a thick peptidoglycan layer, whereas gram-negative bacterial cells are characterized by a thin peptidoglycan layer surrounded by an outer membrane.
- Some prokaryotic cells produce glycocalyx coatings, such as capsules and slime layers, that aid in attachment to surfaces and/or evasion of the host immune system.
- Some prokaryotic cells have fimbriae or pili, filamentous appendages that aid in attachment to surfaces. Pili are also used in the transfer of genetic material between cells.
- Some prokaryotic cells use one or more flagella to move through water. Peritrichous bacteria, which have numerous flagella, use runs and tumbles to move purposefully in the direction of a chemical attractant.
Footnotes
- 1 Y.-H.M. Chan, W.F. Marshall. “Scaling Properties of Cell and Organelle Size.” Organogenesis 6 no. 2 (2010):88–96.
- 2 F. Rothfuss, M Bender, R Conrad. “Survival and Activity of Bacteria in a Deep, Aged Lake Sediment (Lake Constance).” Microbial Ecology 33 no. 1 (1997):69–77.
- 3 R. Sinclair et al. “Persistence of Category A Select Agents in the Environment.” Applied and Environmental Microbiology 74 no. 3 (2008):555–563.
- 4 T.J. Silhavy, D. Kahne, S. Walker. “The Bacterial Cell Envelope.” Cold Spring Harbor Perspectives in Biology 2 no. 5 (2010):a000414.
- 5 B. Zuber et al. “Granular Layer in the Periplasmic Space of Gram-Positive Bacteria and Fine Structures of Enterococcus gallinarum and Streptococcus gordonii Septa Revealed by Cryo-Electron Microscopy of Vitreous Sections.” Journal of Bacteriology 188 no. 18 (2006):6652–6660
- 6 L. Gana, S. Chena, G.J. Jensena. “Molecular Organization of Gram-Negative Peptidoglycan.” Proceedings of the National Academy of Sciences of the United States of America 105 no. 48 (2008):18953–18957.
- 7 J.A. Garnetta et al. “Structural Insights Into the Biogenesis and Biofilm Formation by the Escherichia coli Common Pilus.” Proceedings of the National Academy of Sciences of the United States of America 109 no. 10 (2012):3950–3955.
- 8 T. Proft, E.N. Baker. “Pili in Gram-Negative and Gram-Positive Bacteria—Structure, Assembly and Their Role in Disease.” Cellular and Molecular Life Sciences 66 (2009):613.


