3.2: Bacterial Quorum Sensing, Pathogenicity Islands, and Secretion Systems (Injectosomes)
- Page ID
- 3151
- Define the following:
- pathogenicity
- virulence
- Even though a microorganism may be considered pathogenic, it still may not be able to cause disease upon entering the body. Discuss why.
- Define and briefly describe the overall process of quorum sensing in bacteria and how it may enable bacteria to behave as a multicellular population.
- State at least two possible advantages of individual bacterial behavior.
- State at least two possible advantages of multicellular bacterial behavior.
- State what is meant by intraspecies, interspecies, and interkingdom communication.
- State the function of bacterial secretions systems (injectisomes) such as the type 3 and type 6 secretion systems in bacterial pathogenicity.
In this Learning Object we are going to look at several aspects of bacterial genetics that are directly related to bacterial pathogenicity, namely, quorum sensing, pathogenicity islands, and secretion systems. Pathogenicity and virulence are terms that refer to an organism's ability to cause disease. Pathogenicity is the ability of a microbe to cause disease and inflict damage upon its host, whereas virulence is the degree of pathogenicity within a group or species of microbes as indicated by case fatality rates and/or the ability of the organism to invade the tissues of the host. The pathogenicity of an organism, that is its ability to cause disease, is determined by its virulence factors .
Many of the virulence factors that enable bacteria to colonize the body and/or harm the body are the products of quorum sensing genes. Many bacteria are able to sense their own population density, communicate with each other by way of secreted chemical factors, and behave as a population rather than as individual bacteria . This plays an important role in pathogenicity and survival for many bacteria.
Bacterial Quorum Sensing
Bacteria can behave either as individual single-celled organisms or as multicellular populations. Bacteria exhibit these behaviors by chemically "talking" to one another through a process called quorum sensing. Quorum sensing involves the production, release, and community-wide sensing of molecules called autoinducers that modulate gene expression, and ultimately bacterial behavior, in response to the density of a bacterial population.
To initiate the process of quorum sensing, bacterial genes code for the production of signaling molecules called autoinducers that are released into the bacterium's surrounding environment. These signaling molecules then bind to signaling receptors either on the bacterial surface or in the cytoplasm. When these autoinducers reach a critical, threshold level, they activate bacterial quorum sensing genes that enable the bacteria to behave as a multicellular population rather than as individual single-celled organisms (Figure \(\PageIndex{3}\).2.2). The autoinducer/receptor complex is able to bind to DNA promoters and activate the transcription of quorum sensing-controlled genes in the bacterium. In this way, individual bacteria within a group are able to benefit from the activity of the entire group.
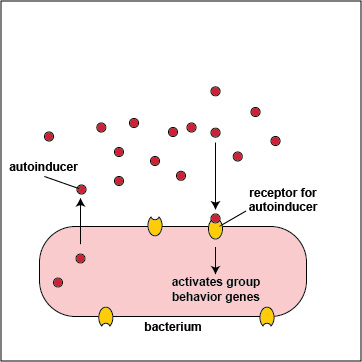
- In Gram-negative bacteria, the autoinducers are typically molecules called acyl-homoserine lactones or AHL. AHLs diffuse readily out of and into bacterial cells where they bind to AHL receptors in the cytoplasm of the bacteria. When a critical level of AHL is reached, the cytoplasmic autoinducer/receptor complex functions as a DNA-binding transcriptional activator.
- In Gram-positive bacteria, the autoinducers are oligopeptides, short peptides typically 8-10 amino acids long. Oligopeptides cannot diffuse in and out of bacteria like AHLs, but rather leave bacteria via specific exporters. They then bind to autoinducer receptors on the surface of the bacterium. When a critical level of oligopeptide is reached, the binding of the oligopeptide to its receptor starts a phosphorylation cascade that activates DNA-binding transcriptional regulatory proteins called response regulators.
The outcomes of bacteria-host interaction are often related to bacterial population density. Bacterial virulence, that is its ability to cause disease, is largely based on the bacterium's ability to produce gene products called virulence factors that enable that bacterium to colonize the host, resist body defenses, and harm the body.
At a low density of bacteria, the autoinducers diffuse away from the bacteria (Figure \(\PageIndex{3}\).2.2). Sufficient quantities of these molecules are unable to bind to the signaling receptors on the bacterial surface and the quorum sensing genes that enable the bacteria to act as a population are not activated. This enables the bacteria to behave as individual, single-celled organisms.
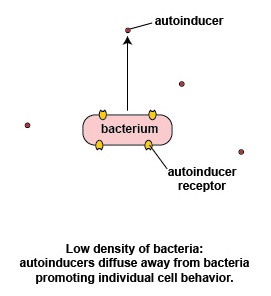
Possible advantages of individual bacterial behavior seen at low bacterial density
If a relatively small number of a specific bacterium were to enter the body and immediately start producing their virulence factors, chances are the body's immune systems would have sufficient time to recognize and counter those virulence factors and remove the bacteria before there was sufficient quantity to cause harm. The bacterium instead utilizes genes that enable it to act as an individual organism rather than as part of a multicellular population.
Acting as individual organisms may better enable that low density of bacteria to gain a better foothold in their new environment in the following ways:
1. Many bacteria are capable of motility and motility serves to keep bacteria in an optimum environment via taxis .
Motility and chemotaxis probably help some intestinal and urinary pathogens to move through the mucous layer so they can attach to the epithelial cells of the mucous membranes. In fact, many bacteria that can colonize the mucous membranes of the bladder and the intestines are motile. Motility probably helps these bacteria move through the mucus in places where it is less viscous.
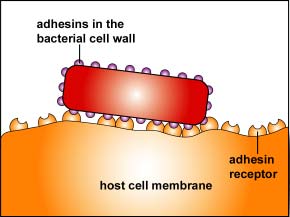
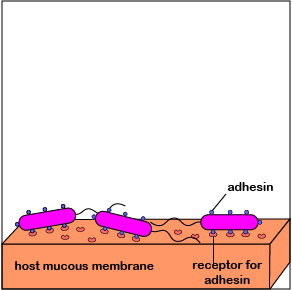
2. One of the body's innate defenses is the ability to physically remove bacteria from the body through such means as the constant shedding of surface epithelial cells from the skin and mucous membranes, the removal of bacteria by such means as coughing, sneezing, vomiting, and diarrhea, and bacterial removal by bodily fluids such as saliva, blood, mucous, and urine. Bacteria may resist this physical removal by producing pili (see Figure \(\PageIndex{3}\)), cell wall adhesin proteins (Figure \(\PageIndex{3}\).2.4), and/or biofilm-producing capsules . Some pili, called type IV pili also allow some bacteria to "walk" or "crawl" along surfaces to spread out and eventually form microcolonies.
Figure \(\PageIndex{3}\).2.3: Adhesive Tip of Bacterial Pili Binding to Host Cell Receptors
3. Many bacteria secrete an extracellular polysaccharide or polypeptide matrix called a capsule or glycocalyx that enables the bacteria to adhere to host cells, resist phagocytosis, and form microcolonies. As the bacteria geometrically increase in number by binary fission, so does the amount of their secreted autoinducers, and production of high levels of autoinducers then enables the population of bacteria to communicate with one another by quorum sensing. At a high density of bacteria, large quantities of autoinducers are produced (Figure \(\PageIndex{3}\).2.5) and are able to bind to the signaling receptors on the bacterial surface in sufficient quantity so as to activate the quorum sensing genes that enable the bacteria to behave as a multicellular population (Figure \(\PageIndex{3}\).2.1).
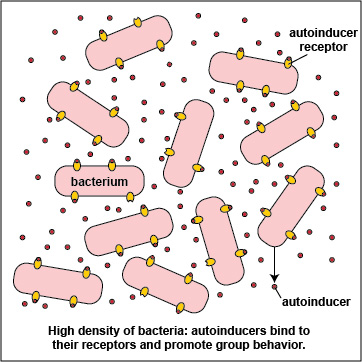
Advantages of Multicellular Behavior seen at High Bacterial Density
- By behaving as a multicellular population, individual bacteria within a group are able to benefit from the activity of the entire group. As the entire population of bacteria simultaneously turn on their virulence genes, the body's immune systems are much less likely to have enough time to counter those virulence factors before harm is done.
- This triggers production of an extracellular adhesive matrix (glycocalyx) enabling the bacteria to form microcolonies and irreversibly attachment to the mucous membranes. Biofilm formation begins.
- Virulence factors such as exoenzymes and toxins can damage host cells enabling the bacteria in the biofilm to obtain nutrients. The biofilm continues to develop and mature.
- As the area becomes over-populated with bacteria, quorum sensing enables some of the bacteria to escape the biofilm, often by again producing flagella, and return to individual single-celled organism behavior in order to find a new sight to colonize.
Pseudomonas aeruginosa is an example of a quorum sensing bacterium. P. aeruginosa causes severe hospital-acquired infections, chronic infections in people with cystic fibrosis, and potentially fatal infections in those who are immunocompromised.
1. When P. aeruginosa first enters the body, they are at a low density of bacteria. The autoinducers diffuse away from the bacteria (Figure \(\PageIndex{3}\).2.2), sufficient quantities of these molecules are unable to bind to the signaling receptors, and the quorum sensing genes that enable the bacteria to act as a population are not activated. The P. aeruginosa continue to function as individual bacteria. Motility genes (coding for flagella) and adhesin genes (coding for pili and cell wall adhesins) are expressed. The flagella enable the initial bacteria to swim through mucus towards host tissues such as mucous membranes. Pili then enable the bacteria to reversibly attach to host cells in order to resist flushing and begin colonization (Figure \(\PageIndex{3}\).2.6; left). Type IV pili, which enable a twitching motility in some bacteria, then enable the bacteria as they replicate to crawl along and spread out over the mucous membranes (Figure \(\PageIndex{3}\).2.6; middle). The pili subsequently retract and bacterial cell wall adhesins enable a more intimate attachment of the bacterium to the mucous membranes (Figure \(\PageIndex{3}\).2.6; right).
|
You Tube movie of motile Pseudomonas
|
2. Once P. aeruginosa has colonized, it is able to replicate geometrically and achieve a high population density. Quorum sensing genes are activated and the bacteria function as a population. This triggers production of an extracellular polysaccharide called alginate to form microcolonies and enables irreversible attachment to the mucous membranes (Figure 3.2.7; left). Biofilm formation begins.
3. Quorum sensing genes coding for enzymes and toxins that damage host cells are produced. These are injected into the host cells by way of an injectosome. This releases nutrients for the bacteria in the biofilm. The bacteria continue to replicate as the biofilm continues to develop, mushroom up, and mature (Figure 3.2.7; middle).
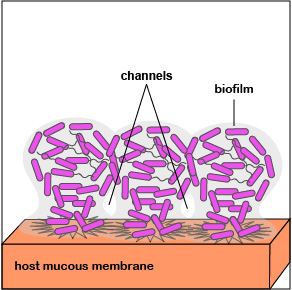
4. As the bacteria replicate, the biofilm continues to mature (Figure 3.2.7; right). Water channels form within the biofilm to deliver water, oxygen, and nutrients to the growing population of P. aeruginosa. The high density of bacteria bacteria are now acting as a multicellular population rather than as individual bacteria.
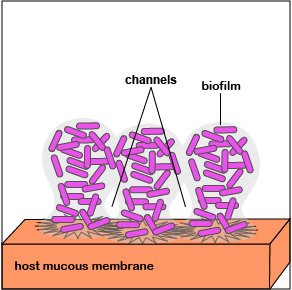
The biofilm enables bacteria to:
- resist attack by antibiotics;
- trap nutrients for bacterial growth and remain in a favorable niche;
- adhere to environmental surfaces and resist flushing;
- live in close association and communicate with other bacteria in the biofilm; and
- resist phagocytosis and attack by the body's complement pathways.
5. When the population of P. aeruginosa begins to outgrow their local environment, quorum sensing enables them to turn off adhesin genes and turn on flagella genes that allow some of the bacteria to spread out of the biofilm to new location within that environment via motility (Figure \(\PageIndex{3}\).2.8).
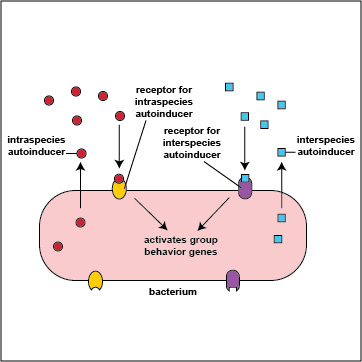
It turns out that bacteria are multilingual. They use quorum sensing not only to "talk" to members their own species (intraspecies communication), but also to "talk" to bacteria that are not of their genus and species (interspecies communication). Intraspecies autoinducers and receptors enable bacteria to communicate with others of their own species while interspecies autoinducers and receptors enable bacteria to communicate with bacteria of a different species or genus (Figure \(\PageIndex{3}\).2.9). The autoinducers for interspecies communications are referred to as AI-2 family autoinducers and are different from the intraspecies (AI-1) autoinducers. In some cases bacteria use interspeciecies communication to work cooperatively with various other bacteria in their biofilm to the benefit all involved; in other cases, bacteria may use interspecies communication in such a way that one group benefits at the expense of another.
Figure \(\PageIndex{3}\).2.9: Intraspecies and Interspecies Communication. Intraspecies autoinducers and receptors enable bacteria to communicate with others of their own species while interspecies autoinducers and receptors enable bacteria to communicate with bacteria of a different species or genus.
Furthermore, bacteria are capable of interkingdom communication, communication between bacteria and their animal or plant host. Increasing numbers of bacteria are being found that have signaling receptors that recognize human hormones. For example, a number of bacteria that are pathogens of the human intestinal tract have a sensing molecule called QseC that binds the human hormones adrenaline and noradrenaline. This, in turn, activates various virulence genes of the bacteria. On the other hand, some bacterial autoinducers can enter human host cells and regulate human cellular function. For example, at low concentration some bacterial autoinducers suppress host immune responses thus better enabling those bacteria to better establish themselves in the body. At high concentrations, however, they stimulate an inflammatory response in the host to help the bacteria to spread from the initial infection site. One bacterial autoinducer has been found to initiate apoptosis (cell suicide) in phagocytes such as neutrophils and macrophages.
Bacterial Pathogenicity Islands
The genomes of pathogenic bacteria, when compared with those of similar nonpathogenic species or strains, often show extra genes coding for virulence factors , that is, molecules expressed and secreted by the bacterium that enable them to colonize the host, evade or inhibit the immune responses of the host, enter into or out of a host cell, and/or obtain nutrition from the host. These include virulence factors such as capsules, adhesins, type 3 secretion systems, invasins, and toxins.
Most genes coding for virulence factors in bacteria are located in pathogenicity islands or PAIs and are usually acquired by horizontal gene transfer . These PAIs may be located in the bacterial chromosome, in plasmids, or even in bacteriophage genomes that have entered the bacterium. The genomes of most pathogenic bacteria typically contain multiple PAIs that can account for up to 10 transpoases ,- 20% of the bacterium's genome. PAIs carry genes such as integrases , or insertion sequences that enable them to insert into host bacterial DNA. Transfer RNA (tRNA) genes are often the target site for integration of PAIs. Conjugative plasmids are the most frequent means of transfer of PAIs from one bacterium to another and the transfer of PAIs can then confer virulence to a previously nonpathogenic bacterium.
Type 3 Secretion Systems (T3SS or Injectisomes) and Type 6 Secretion Systems (T6SS)
Many bacteria involved in infection have the ability to co-opt the functions of host cells for the bacterium’s own benefit. This is done by way of bacterial secretions systems that enable the bacterium to directly inject bacterial effector molecules into the cytoplasm of the host cell in order to alter its cellular machinery or cellular communication to the benefit of the bacteria.
The most common type is the type 3 secretion system or T3SS (Figure \(\PageIndex{3}\).2.10). A secretion apparatus in the cytoplasmic membrane and cell wall of the bacterium polymerizes a hollow needle that is lowered to the cytoplasmic membrane of the host cell and a translocon protein is then delivered to anchor the needle to the host cell. Effector proteins in the bacterium can now be injected into the cytoplasm of the host cell. The delivery system is sometimes called an injectisome. (A type 4 secretion system can transfer effector proteins and/or DNA into the host cell because it is similar to the conjugation transfer system initiated by tra genes discussed under horizontal gene transfer.)
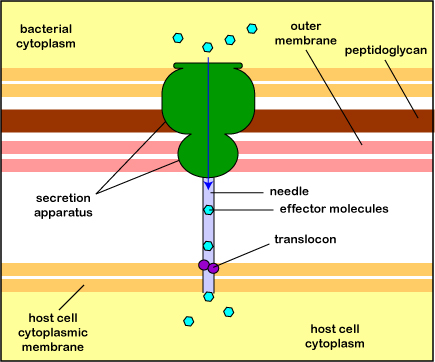
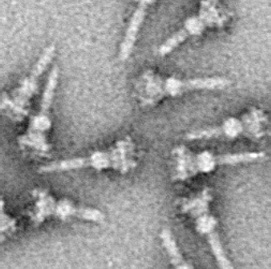
Electron micrograph of an injectisome. A transmission electron-microscope image of isolated T3SS needle complexes from Salmonella typhimurium. (CC BY-SA 2.5; Schraidt O, Lefebre MD, Brunner MJ, Schmied WH, Schmidt A, Radics J, Mechtler K, Galán JE, Marlovits TC - Cropped image from Schraidt et al. (2010), Topology and Organization of the Salmonella typhimurium Type III Secretion Needle Complex Components. PLoS Pathog 6(4): e1000824.doi:10.1371/journal.ppat.1000824)
Some bacteria, such as Pseudomonas aeruginosa and Vibrio cholerae, produce a type 6 secretion system, or T6SS, that consists of a protein tube surrounded by a contractile sheath, similar to the tail of T4-bacteriophages (a bacteriophage is a virus that only infects bacteria.) The type 6 secretion system not only injects effector molecules into eukaryotic cells, but also is able to inject antibacterial effector molecules into other bacteria in order to kill those bacteria. Predator bacteria can use their T6SS to kill prey bacteria. In fact, V. cholerae and P. aeruginosa have been shown to "duel" with one another via their respective T6SSs.
V. cholerae also uses its T6SS to promote horizontal gene transfer by way of transformation. Individual V. cholerae cells also use their T6SS to attack one another upon cell-to-cell contact. Most members of the population, however, produce immunity proteins that protect them from being killed by the effector molecules that are injected. Not all strains of V. cholerae in the population, however, produce these immunity proteins and these non-immune cells are subsequently lysed, releasing their DNA into the environment. This DNA can then be taken up by neighboring competent V. cholerae via transformation.
- Briefly describe how bacterial quorum sensing may play a role in pathogenicity by:
- Promoting colonization of a new host by bacteria that have just entered the body.
- Enabling the bacterium to persist within that host once they have colonized.
- Allowing some of the bacteria to spread to a new location within a host or to a new host.
- Briefly describe how the ability to produce a type 3 secretion system might play a role in a pathogen colonizing the body and causing an infection.
Summary
- Pathogenicity is the ability of a microbe to cause disease and inflict damage upon its host; virulence is the degree of pathogenicity within a group or species of microbes.
- The pathogenicity of an organism is determined by its virulence factors.
- Virulence factors enable that bacterium to colonize the host, resist body defenses, and harm the body.
- Most of the virulence factors are the products of quorum sensing genes.
- Quorum sensing involves the production, release, and community-wide sensing of molecules called autoinducers that modulate gene expression, and ultimately bacterial behavior, in response to the density of a bacterial population.
- The outcomes of bacteria-host interaction are often related to bacterial population density.
- At a low density of bacteria, the autoinducers diffuse away from the bacteria and there are insufficient quantities of these molecules to activate the quorum sensing genes that enable the bacteria to act as a population. As a result the bacteria behave as individual, single-celled organisms.
- Acting as individual organisms may enable a low density of bacteria to gain a better foothold in their new environment by enabling bacteria to use motility and taxis to contact host cells, use pili to initially adhere to and crawl over host cell surfaces, use adhesins to adhere to host cells and resist flushing, and secrete a glycocalyx to form microcolonies.
- As the bacteria increase in numbers geometrically as a result of binary fission and reach high density, large quantities of autoinducers are produced and are able to bind to the signaling receptors on the bacterial surface in sufficient quantity so as to activate the quorum sensing genes that enable the bacteria to now behave as a multicellular population.
- By behaving as a multicellular population, individual bacteria within a group are able to benefit from the activity of the entire group.
- As the entire population of bacteria simultaneously turn on their virulence genes, the body's immune systems are much less likely to have enough time to counter those virulence factors before harm is done. Virulence factors such as exoenzymes and toxins can damage host cells enabling the bacteria in the biofilm to obtain nutrients.
- As the area becomes over-populated with bacteria, quorum sensing enables some of the bacteria to escape the biofilm and return to individual single-celled organism behavior in order to find a new sight to colonize.
- Quorum sensing enables bacteria to communicate with members of their own species, with other species of bacteria, and with their eukaryotic host cells.
- Most genes coding for virulence factors in bacteria are located in pathogenicity islands or PAIs and are usually acquired by horizontal gene transfer.
- Many bacteria involved in infection have the ability to co-opt the functions of the host cell for the bacterium’s own benefit by producing secretions systems that enable the bacterium to directly inject bacterial effector molecules into the cytoplasm of the host cell in order to alter the host cell’s cellular machinery, cellular function, or cellular communication.