1.10: Genetic Pathways
- Page ID
- 73675
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Understand how multiple genes often function together to control a trait
- Understand the concept of a biosynthetic pathway
- Understand the concept of a regulatory pathway
- Understand the concept of a network
- Understand how mutations in different genes of a pathway can cause the same phenotype
- Understand how mutations in different genes of a pathway might cause different phenotypes
Introduction
Genes do not function in isolation but rather suites of genes act in concert to perform biological functions. When different genes function in different sequential steps of a biological process, this is known as a genetic pathway. Perhaps the most conceptually intuitive type of pathway is a biosynthetic pathway, where a precursor molecule is chemically modified through a series of enzymatically catalyzed intermediate steps to produce a bioactive product. Since enzymes are (proteins), the genes that encode these enzymes are considered a genetic pathway. In a regulatory pathway, some type of stimulus leads to a change in the expression or activity of a particular gene product, which in turn acts to alter the expression or activity of another gene product or products, which in turn could regulate yet another level of activity.
Ultimately, these regulatory changes in expression or activity lead to a response to the stimulus, typically involving changes in gene expression. Again, the genes that encode the various types of regulatory molecules, mainly proteins, constitute a genetic pathway. Pathways are often targets in the biotechnological manipulation of traits, which requires an understanding of how pathways behave. Also note that many biosynthetic and regulatory systems are better described as networks because of often, pathways branch or converge and interact with other systems. However, for simplicity such complex networks will not be discussed here.
Biosynthetic pathways
Genes encode enzymes that catalyze steps in the synthesis of a compound. A biosynthetic pathway actually describes a process of converting a precursor substrate into a product. In addition to the precursor substrate and product, it includes the enzymes, the chemical reactions catalyzed by the enzymes and all the intermediate compounds. A pathway consists of a series of steps where a precursor molecule acts as a substrate for an enzyme, which catalyzes a chemical reaction to produce a product, which then serves as a substrate for a subsequent step.

We have already discussed the one gene, one enzyme hypothesis which was an important advance in the discovery of how genes encode proteins. This analysis also provides a nice example of the concept of a biosynthetic pathway. This hypothesis was discovered by analyzing auxotrophic neurospora mutants that were defective in the production of amino acids. That is, the biosynthetic pathways producing particular amino acids were disrupted by mutations. The text discusses the analysis of the arginine pathway. Mutants that disrupt this pathway cannot grow on minimal medium lacking arginine but can grow if arginine is supplied in the medium. The same fundamental pathway also produces arginine in plants.
Mutations that disrupt the function of particular enzymes in the pathway block the progression of the pathway at that corresponding step. For example, a mutation in the argF gene would disrupt the enzyme ornithine carbamoyltransferase (OTC). This would block step 6 in the pathway, the conversion of L-ornithine to L-citruline. As a result, the intermediate compound L-Ornithine will likely accumulate, whereas all the compounds that occur later in the pathway (i.e., L-Citruline, L-argininosuccinate and L-arginine) will be deficient.
The anthocyanin pathway
The anthocyanin pathway is a slightly more complex pathway, although still simple as far as these things go. It is of significant interest to horticulturists because anthocyanin pigments are responsible for many plant colors, particularly in flowers and fruits (think red wine). Anthocyanin pigments range from orange to red, purple, and blue. In addition to the attractive colors, anthocyanins provide, there is also considerable recent interest in anthocyanins for their dietary value, acting as strong antioxidants and possibly anti-cancer agents.
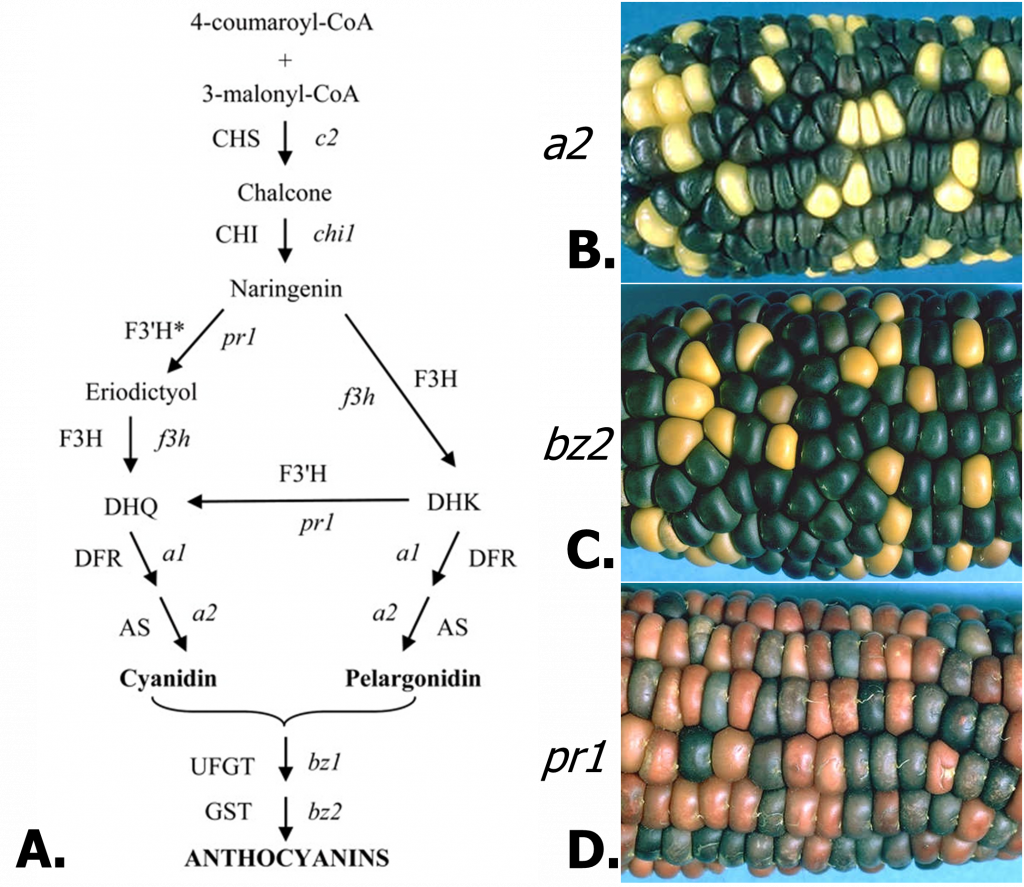
Effects of mutations at different steps in anthocyanin biosynthesis.
Of course, each enzyme in the anthocyanin pathway is encoded by a gene. As such, mutations can generate variation or disrupt the pathway. The maize anthocyanin pathway and examples of mutants are shown in Figure 2. The a2 mutant causes a deficiency in the anthocyanidin synthase enzyme, blocking the production of any pigmented compounds. The a1 and c2 mutants are similarly colorless. The bz2 mutant is deficient in an enzyme related to glutathione-S-transferase, which is required for transport of anthocyanidins into the vacuole, resulting in the bronze-colored pigment due to the more alkaline environment of the cytosol. The bz1 mutant (not shown) produces a similar phenotype. Mutation of the pr1 gene eliminates just one class of anthocyanins. The pr1 gene encodes flavanone 3′-hydroxylase, required for the production of cyanidin, which generates purple anthocyanin. In the mutant, only pelargonidin is synthesized, producing a red pigment.
Regulatory pathways
A regulatory pathway refers to a situation where one gene or gene product controls the expression or activity of another gene or gene product, which may in turn also perform a regulatory function. As we discussed, gene function can be regulated at many points from transcriptional regulation to post-translational regulation. We will examine a couple of specific examples of regulatory pathways but first let’s consider some of the general features of regulatory pathways. Understanding the logic of regulatory pathways will become important later when we seek to use biotechnology to manipulate traits by altering their regulation.
A simplistic but useful way to help understand the logic of regulatory pathways is to consider them as a series of switches with on and off states.
Positive vs. negative regulators
A key feature of regulatory pathways is that regulators might function either positively or negatively. A positive regulator is one that functions to activate the next component in the pathway, whereas a negative regulator would inhibit the next component. For example, a transcription factor that activated the transcription of a gene would be considered a positive regulator, while one that repressed the transcription of a gene would be a negative regulator. Another common type of regulator is a protein kinase. Protein kinases are proteins that add a phosphate group to another protein. Phosphorylation is a common type of post-translational modification that can regulate the activity of proteins; sometimes the phosphorylated protein becomes activated and sometimes it becomes repressed.
A positive regulatory step is represented with an arrow. For example, “protein A activates protein B” would be represented like:
\[\text{A} \to \text{B}\]
A negative regulatory step is represented with a bar. “Protein C represses protein D” would be represented like:
\[\text{C} \dashv \text{D}\]
Active vs. inactive states of regulators
A second point to consider in the logic of regulatory pathways is the activity state of each component. A useful way to think about this is that each component can have an “on” or an “off” state. Of course, in reality, many proteins can have intermediate levels of activity but for the sake of simplicity, we will just consider the on and off states.
The effect of a regulator being in the on or off state depends on whether it is a positive or negative regulator.
If a positive regulator is in the on state, it will function to activate the next factor. In the example of 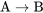 , if A is in the on or active state, it will function to switch B to the on or active state.
, if A is in the on or active state, it will function to switch B to the on or active state.
If a negative regulator is in the on state, it will function to switch the next factor to the off or inactive state. For 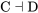 , if C is in the on or active state, it will switch D to the off or inactive state.
, if C is in the on or active state, it will switch D to the off or inactive state.
Regulatory pathways sometimes contain many, sometimes few, steps. They can also contain a mixture of positive and negative regulators. Let us explore a few hypothetical examples to see how the pieces fit together to regulate plant responses to stimuli.
When a pathway is depicted, all the steps are shown. For the sake of simplicity, it is assumed that the default activity state of any given component is such that the preceding step functions to change it. We will begin with a simple example containing several positive regulatory steps that activate a cellular activity, Activity 1, in response to Stimulus 1. In the pathway shown below, the Stimulus 1 activates A. Therefore, we assume that in the absence of this stimulus, A is in the inactive or off state. Since A is inactive, it is not functioning to activate B, which is therefore also in the inactive state, and likewise for C. Ultimately Activity 1 does not occur in the absence of Stimulus 1 but does occur in response to the stimulus.
Stimulus 1 → A → B → C → Activity 1
We can use color coding to help visualize this. Light grey represents the OFF state and blue represents the ON state.
No Stimulus 1 → A → B → C → Activity 1 (OFF)
Stimulus 1 → A → B → C → Activity 1 (ON)
Or we can make a table to help keep track of the states of each component in the presence or absence of stimulus.
| Stimulus 1 → | A → | B → | C → | Activity 1 |
| NO | OFF | OFF | OFF | NO |
| YES | ON | ON | ON | YES |
Now let us look at another example containing both positive and negative regulators.
Stimulus 2 → D → E 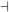 F → Activity 2
F → Activity 2
In this case, components D and E would be in the inactive states in the absence of stimulus, because they would not have been activated. But E functions as a negative regulator of F. Since E is inactive, it is not functioning to inhibit F, which then remains in the active state promote cellular Activity 2. When Stimulus 2 is present, D and E become activated, and E functions to inactivate F. Since F is off, Activity 2 does not occur. Thus, the net response to Stimulus 2 is the repression of Activity 2.
\[\dashv\]
\[\dashv\]
OR
| Stimulus 2 → | D → | E  |
F → | Activity 2 |
| NO | OFF | OFF | ON | YES |
| YES | ON | ON | OFF | NO |
Now that we have considered regulatory pathways from a hypothetical perspective, let’s consider a couple real ones. As mentioned, regulatory pathways can take on many forms. They can consist of a series of transcription factors that regulate the expression of one another’s genes, ultimately resulting in the regulation of genes that effect some biological response. Here, we will look at a signal transduction pathway consisting of a series of post-translational modifications that occur response to a hormone stimulus and culminate in gene expression changes to effect a response.
Signal transduction pathway for hormone signaling
Brassinosteroids (BRs) are a class of plant steroid hormones that control many aspect of plant physiology. They are most noted for their role in promoting plant growth and mutants deficient in BR biosynthesis or signaling show dwarfism. BRs also promote a range of other responses, including yield and increased stress resistance.
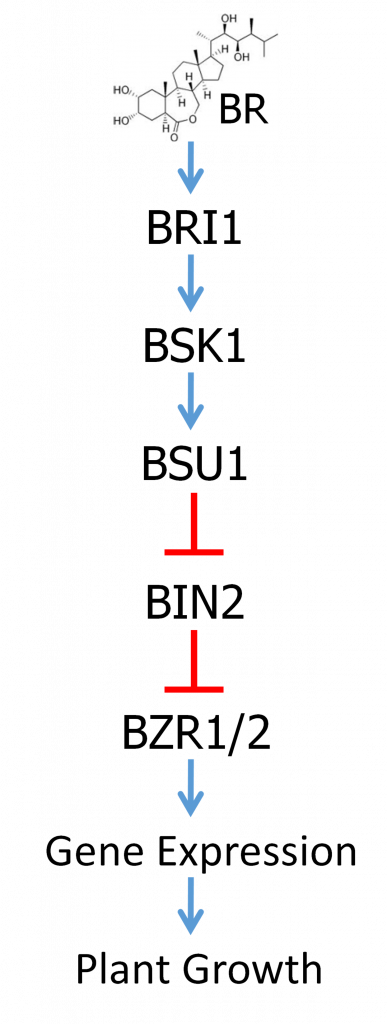
BR hormones, of which brassinolide (BL) is considered the most active, are recognized by a receptor called BRI1. BRI1 is a type of receptor known as a receptor kinase, which spans the cell membrane. On the outside of the cell is a receptor domain which specifically recognizes and binds to BR. There is also a membrane-spanning domain and then a protein kinase domain inside the cell. Binding to BR outside the cell then activates the protein kinase domain inside the cell. Protein kinases are enzymes that function to add phosphate groups to other proteins. As previously discussed, phosphorylation can alter the activity of a protein. Activation of the BRI1 receptor kinase then results in the activation of a signaling pathway that ultimately regulates the activity of transcription factors inside the nucleus, changing the expression of genes that regulate growth and stress resistance.
The BR signal transduction pathway is well studied and is summarized in Figure 3. Do not be concerned with memorizing the names of all these factors, but just focus on understanding the regulatory logic. The overall regulatory relationships of the pathway are depicted in Figure 3. The main target of regulation is a pair of closely related transcription factors called BZR1 and BZR2. BZR1/2 are negatively regulated by a protein called BIN2 in the absence of BR. BIN2 is a protein kinase that phosphorylates BZR1/2, which results in BZR1/2’s exclusion from the nucleus and their proteolytic degradation. In the absence of BR, BIN2 contains a phosphate group that is required for its activity.
Binding of BR to the BRI1 receptor domain outside the cell results in the activation of the kinase domain inside the cell. Activated BRI1 then phosphorylates a protein called BSK. Phosphorylated BSK binds yet another protein called BSU1, which is a protein phosphatase. Protein phosphatases are enzymes that remove phosphate groups from other proteins. BSK binding activates BSU1 which then catalyzes the removal of the phosphate on BIN2, thereby inactivating BIN2. Thus, BSU1 is a negative regulator of BIN2. When BIN2 is inactivated, that allows BZR1/2 to accumulate, to enter the nucleus, and to regulate the expression of genes that effect the BR response, including promoting plant growth and yield.
Effects of mutation on different steps
Mutations can affect regulatory pathways in multiple ways. Most mutations are loss-of-function mutations where the function of the gene or gene product is partly or completely impaired. Considering the BR signaling pathway, the phenotypes of mutation will differ depending on whether the mutation affects a positive or negative regulator of the pathway. A loss-of-function mutation in the bri1 gene will block the perception of the hormone and therefore the pathway will not be activated; a dwarf phenotype will result. On the other hand, a loss-of-function in the bin2 gene will release BZR1/2 from regulation. The result will be constant BZR1/2 activity regardless of whether BR hormone is present. This might be expected to generate giant plants but in fact the effects of deregulating hormone responses are more complex—let us just call it unregulated growth.
Mutations can also be gain-of-function, where the gene product assumes a form that is constantly in the active state. Such mutations are typically dominant and are much less common than loss-of-function. Again, the outcome of a gain-of-function mutation on the BR signaling pathway will vary depending on whether it affects a positive or negative regulator of the pathway. A gain-of-function mutation in a positive regulator would cause deregulated hormone responses, whereas such a mutation in a negative regulator would permanently shut down the pathway and cause dwarfism.
Complementary gene action and genetic epistasis in the context of pathways
Considering that biological processes are controlled by multiple genes acting together, several genetic principles make more sense. First, it becomes clear how mutations in different genes can produce the same phenotypic effects. Since multiple genes are often required for a single biological process, disruption of different genes in the process might have the same net effect. From this follows the concept of complementary gene action. Mutant plants with the same phenotype are crossed together and the offspring is normal looking. The mutations in the parent plants likely affected different genes in the same pathway. Finally, the concept of epistasis can be best understood in the context of pathways. Note that the term epistasis is used differently by different geneticists. In quantitative genetics, epistasis refers to any gene interaction. Here, the term epistasis refers to a specific type of gene interaction where the action of one gene is masked by the action of another gene. In other words, if two mutations are combined in an individual, only one of the phenotypes is apparent. The mutant whose phenotype is apparent is said to be epistatic to the one that is masked.
Complementary gene action in a regulatory pathway
The concept of complementary gene action is no different in a regulatory pathway than in a biosynthetic pathway. Mutations in different genes that have similar roles in the pathway (e.g., are both positive regulators of the pathway) would be expected to cause similar mutant phenotypes. If such mutants in different genes were crossed together, the F1 progeny would be normal because both genes would be heterozygous and therefore a functional copy of both factors would be present.
Epistasis in a regulatory pathway
Epistasis can be quite complex, especially in regulatory pathways. Unlike complementary gene action, epistasis is not restricted to recessive loss-of-function mutations. The situation is further complicated by the presence of positive and negative regulators in a pathway. In contrast to a biosynthetic pathway, the rule of thumb for a regulatory pathway is that the more “downstream” gene is epistatic to the more “upstream” one. Let’s turn again to the BR signaling pathway and look at a couple of examples to understand why.
First consider loss-of-function mutations in the bri1 and bin2 genes, which in single mutants cause a dwarf phenotype and an unregulated growth phenotype, respectively. If the two mutations are combined into a double mutant plant, what do we expect? The bri1 mutations disrupt the receptor function required to initiate signaling through the pathway. However, BIN2 functions to inhibit the activity of pathway by repressing the function of BZR1/2. If BIN2 is rendered non-functional, then BZR1/2 will be active and promote the growth response. This will be true even if previous steps are blocked by the bri1 mutation. So the double mutant would show the bin2 unregulated growth phenotype making bin2 epistatic to bri1.
What about a gain-of-function mutation in BZR1 or BZR2 combined with a loss of function mutation in bri1? A gain-of-function mutation would render BZR1/2 constitutively active, regardless of the activity state of BIN2, resulting in the unregulated growth phenotype. Since BIN2 activity would be irrelevant, upstream activities would also be irrelevant. Thus, the double mutant would show unregulated growth and BZR1/2 would be epistatic to bri1.
We saw that genes do not act alone but function in concert with other genes to perform various biological functions such as the biosynthesis of a compound or the regulation of a response to a stimulus. When genes control a sequential series of steps, that is called a pathway. But pathways typically branch and intersect with other pathways to form networks. Mutations affect the activity of pathways in various ways, depending on the nature of the mutation (loss- vs. gain-of-function) and the function of the gene product (positive vs. negative effector).
When mutations affect different steps in a pathway, they can often have a similar overall effect on the pathway and therefore lead to similar phenotypic consequences. Such mutations will complement one another even though their phenotypes are identical. It is also possible that mutations in different steps of a pathway can cause different phenotypes. When such mutations are combined into a double mutant individual, the phenotype of one is often masked by the phenotype of the other epistatic mutation. When pathways are linear, the “upstream” mutation is generally epistatic in biosynthetic pathways and in regulatory pathways it is generally the “downstream” mutation that is epistatic. In branched pathways or networks, it is more difficult to generalize.
Complementary gene action is important for geneticists trying to identify all the steps in a pathway. This is the basis of the classic complementation test used to determine if mutations are allelic or affect independent genes. Epistasis is also useful to geneticists who want to understand whether mutations with different phenotypes affect the same pathway, and if so, to determine the order of gene action.
When biotechnology is used to manipulate a trait, it is really the activity of a pathway or network that is being targeted. The same genetic principles outlined in this section are used to design biotechnological strategies to alter pathway activities so as to produce the desired outcome on the trait of interest.
To answer the following questions, refer to the pathways illustrated in Figure 3. They are labeled slightly differently but a1 encodes DFR, which catalyzes the production of leucoanthocyanidins and a2 encodes AS or LDOX/ANS, which subsequently catalyzes the production of anthocyanidins. Assume that normal, wild-type maize kernels are the deep purple/black color as in Figure 3.
- What would you predict happens to the levels of leucoanthocyanidins in an a2 mutant compared to normal?
- no change
- increase
- decrease
- can’t predict
Answer: b or d.
The expectation would be b, that leucoanthocyanidins would increase. Because the pathway is blocked at the subsequent step, leucoanthocyanidins cannot be converted to anthocyanidins. But since all the preceding enzymes remain functional, those reactions would be expected to continue resulting in the accumulation of leucoanthocyanidins.
d is also a correct response. Biosynthetic pathways do not always behave as expected. For example, there could be feedback inhibition of one or more enzymes by intermediate compounds. Bottom line, we won’t know for certain until the levels are measured experimentally.
- What would you predict happens to the levels of pelargonidin in an pr1 mutant compared to normal?
- no change
- increase
- decrease
- can’t predict
Answer: b or d.
Again, the expectation would be b, that pelargonidin would increase. Normally, some fraction of the intermediate compounds are channeled through the pathway branch leading to cyaniding. Because the pathway leading to cyanidin is blocked it might be expected that all the intermediates would be converted to pelargonidin, causing an increased accumulation.
d is also a correct response. As discussed in problem 1, biosynthetic pathways do not always behave as expected. Feedback inhibition could be a factor or it may be possible that the enzymatic capacity of the pelargonidin branch of the pathway is already saturated. To increase flux through the pathway, it could be required to increase the levels or activities of one or more rate-limiting enzymes. Again, we won’t know for certain until the levels are measured experimentally.
Predict the outcomes of the following mutations on the signaling output, or plant response for the BR signaling pathway.
- A gain-of-function mutation in the BZR2 gene
- increased output
- no change
- decreased output
- can’t tell
Answer: a. Since BZR2 is a positive regulator of BR signaling, increased output from the pathway would be expected from a gain-of-function mutation.
- A loss-of-function mutation in the bsu1 gene. (Study the pathway and think about this one!)
- increased output
- no change
- decreased output
- can’t tell
Answer: c. The output from the pathway would decrease causing decreased plant growth. Even though BSU1 is a negative regulator, its target is BIN2 which is another negative regulator. Without BSU1 function, BIN2 will always be in the ON state and inhibit the activities of BZR1/2, therefore decreasing the response to BRs.
Activity 3
1. Predict the phenotype of a BSK gain-of-function.
- unregulated growth
- no effect
- dwarf
- can’t predict
Answer: a. BSK is a positive regulator of the pathway that functions to activate BSU1. Therefore a gain-of-function would cause it to positively activate the pathway, even in the absence of BR hormone, causing unregulated growth.
2. Now predict the phenotype of a double mutant between the BSK gain-of-function and a BIN2 gain-of-function.
- unregulated growth
- no effect
- dwarf
- can’t predict
Answer: c. The gain-of-function BIN2 mutation locks BIN2 in the ON state where it would function to inhibit the activity of BZR1/2. It would be impervious to regulation by the upstream BSU1, so even if BSU1 were continually active due to the gain-of-function BSK, the BIN2 mutant would inhibit growth and therefore be epistatic.
Now you try a couple. Fill in each of the following tables to determine whether the hypothetical Activity will be activated or repressed in response to each Stimulus.
References
Sharma, M., Cortes-Cruz, M., Ahern, K.R., McMullen, M., Brutnell, T.P., and Chopra, S. (2011). Identification of the Pr1 Gene Product Completes the Anthocyanin Biosynthesis Pathway of Maize. Genetics 188, 69-79.


