1.10: Enzyme Function
- Page ID
- 36752
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Learning Objectives
Goals:
- Carryout a colormetric assay to monitor amylase activity.
STUDENT LEARNING OUTCOMES:
Upon completion of this lab, students will be able to:
- Describe how an enzyme works.
- Explain the effect temperature has on the rate of chemical reactions.
- Explain the effect of pH on enzyme activity.
Introduction
Living organisms sustain the activities of life by carrying out thousands of chemical reactions each minute. These reactions do not occur randomly, but are controlled by biological catalysts called enzymes. Enzymes accelerate the rate of chemical reactions by lowering the activation energy needed to trigger the reaction. Without enzymes, chemical reactions would not occur fast enough to support life. Enzymes are typically proteins and each is composed of a specific sequence of amino acids. Hydrogen bonds form between specific amino acids and help create the 3-dimensional shape that is unique to each enzyme. The shape of an enzyme, particularly its active site, dictates catalytic specificity of a particular enzyme. Each enzyme will only bind with specific molecules, as these molecules must fit with the active site on the enzyme like a lock and key.
A molecule that binds with an enzyme and undergoes chemical rearrangement is called a substrate. The enzyme “E” combines with the substrate molecule(s) “S” at the active site and forms a temporary enzyme-substrate complex “ES”, where the specific reaction occurs. The modified substrate molecule is the product “P” of the reaction. The product separates from the enzyme and is then used by the cell or body. The enzyme is neither consumed nor altered by the reaction and can be used in other catalytic reactions as long as additional substrate molecules are available.
\[\ce{E + S -> ES -> E + P}\nonumber\]
An individual enzyme molecule may facilitate several thousand catalytic reactions per second, and therefore only a small amount of enzyme is needed to transform large amounts of substrate molecules into product. The amount of a particular enzyme found in a cell at any given time is relative to the rate at which the enzyme is being synthesized compared to the rate at which it is degraded. If no enzyme is present, the chemical reaction catalyzed by that enzyme will not occur at a functional rate. However, when the concentration of the enzyme increases, the rate of the catalytic reaction will increase as long as the substrate molecules are accessible.
Various factors can inactivate or denature enzymes by altering their 3-dimensional shape and inhibiting their substrate binding efficiency. Many enzymes function best within a narrow temperature and pH range as substantial changes in temperature or pH disrupt their hydrogen bonds and alter their shape. Change in enzyme shape typically alters the shape of the active site, and affects its ability to bind with substrate molecules. It is the unique structural bonding pattern of an enzyme that determines its sensitivity to change in temperature and pH.
In the following exercise you will explore the effects of pH and temperature on the activity of the enzyme amylase. Amylase is found in the saliva of humans and other animals that consume starch as part of their diet. Starch is a plant polysaccharide composed of many glucose molecules bonded together. Amylase controls the initial digestion of starch by breaking it down into disaccharide maltose molecules. Maltose is ultimately broken down into glucose molecules in the small intestine when other enzymes are utilized.
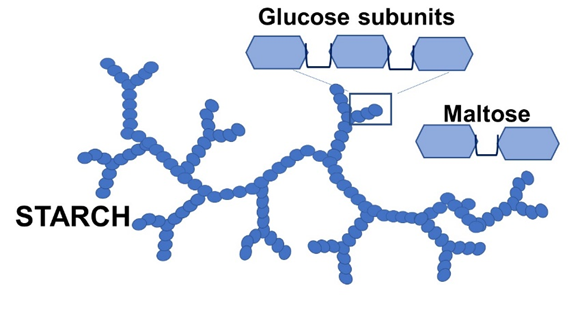
The rate at which starch is digested into maltose is a quantitative measurement of the enzymatic reaction. The rate of starch degradation is relative to the rate at which maltose is produced, however it is easier to test for the presence of starch than it is to measure the rate of maltose production. I2KI will be used as indicator for the presence of starch. When starch is present, I2KI turns a blue-black color. In the presence of maltose, I2KI will not react and remains an amber color.
\[ Starch + I_2KI \rightarrow \text{Dark Blue-Black color} \nonumber\]
\[ Maltose + I_2KI \rightarrow \text{Amber color} \nonumber\]
Your group will be assigned to conduct one or more of the following activities (in part or whole):
I. The effect of pH on amylase activity
II. The effect of temperature on amylase activity
Results from each exercise will be presented to the class and students will be responsible for the information and results from all exercises.
Part I: The Effect of pH on Amylase Activity
Each enzyme has an optimal pH at which it is the most active or effective. A change in pH can alter the bonds of the 3-dimensional shape of an enzyme and cause the enzyme to change shape, which may slow or prohibit binding of the substrate to the active site. You will determine how pH affects amylase activity in this exercise.
Before beginning this experiment, formulate a hypothesis you wish to test and a prediction to evaluate your hypothesis by and write these into the data sheet at the end of the exercise.
Materials
- Test tubes
- Buffers (pH: 1.0, 5.0, 10.0)
- Micropipettes
- Sterile pipette tips
- 1% starch solution
- I2KI solution
- Well plate
- 0.5% amylase solution
- Timer
Procedure
- Label 3 test tubes #1 - #3. Beginning with tube #1 and pH 1, mark one tube with each of the following buffer pH: 1.0, 5.0, and 10.0 (See Table 1 below). After you mark the test tubes, use a P-1000 micropipette and add 4.0 mL of the appropriate buffer to each test tube (4.0 mL pH 1.0 buffer to tube #1, 4.0 mL of pH 5.0 buffer to tube #2, etc). Be sure to place a new tip on the micropipette for each buffer.

Figure 2. Well plates - Using a P-1000 micropipette, add 2.0 mL of the 1% starch solution to each tube and mix by gently swirling the tube and tapping the bottom of the tube against your palm.
- Place 2 drops of I2KI into the compartments of several rows of the test plate so that 24 compartments have clear amber liquid in them.
- Starting with Tube #1 only for now, do the following:
- Using a clean new tip on a P-200 micropipette, draw 50.0 microliters of liquid from the test tube and dispense it into the first compartment of the test plate containing I2KI. The mixture should turn dark blue or black. (This confirms that starch is present in your test tube before the enzyme is added to the tube.)
- Do not touch the I2KI with the Pipette Tip!
- If you do, eject the tip and change to a new clean one.
- Add 400.0 µL of 0.5% amylase solution using a P-1000 micropipette. Begin recording the time in seconds the moment the amylase is added. Mix the tube contents by swirling the tube and gently tapping the bottom of the tube against your palm. Proceed to the next step immediately (within 20 seconds).
- At exactly 20 seconds, transfer 50.0 µL (using your P-200 micropipette) of the reaction mixture from tube #1 into the next new compartment containing I2KI on the test plate.
- Do not touch the I2KI with the Pipette Tip!
- You can continue using the same tip for step 8 below as long as it remains uncontaminated.
- Repeat step 7 every 20 seconds using the next new compartment on the test plate. Continue until a blue-black color is no longer produced and the I2KI solution remains amber (indicating that no starch remains). Then count the total number of compartments that did change colors plus on that did not change colors and multiply by 20. Record the time required for the complete digestion of the starch in Table 1.
If the color of the I2KI continues to change to a darker color after a total of 8 minutes of testing, stop testing that reaction mixture, record 480 seconds as the time in the data table, and proceed with step 9.
- Repeat steps 5 - 8 using tube #2 then #3. You may need to clean your test plate by rinsing it with DI water, tapping it dry, and then adding fresh I2KI to the compartments.
|
Tube |
pH |
Time of Starch Digestion (sec) |
|---|---|---|
|
1 |
1 |
|
|
2 |
5 |
|
|
3 |
10 |
Data Analysis
1. How would you interpret the results shown in Table 1?
Part II: The Effect of Temperature on Amylase Activity
Chemical reactions speed up as temperature increases. A 10o C rise in temperature typically results in a two- to threefold increase in the rate of reaction. However, at high temperatures proteins can be irreversibly denatured and substrate binding is prohibited. The activity of an enzyme is dependent on its proper structure, and the optimum temperature for activity may vary depending on the structure of the enzyme.
Before beginning this experiment, formulate a hypothesis you wish to test and a prediction that can be used to evaluate your hypothesis. Write these into the data sheet at the end of the exercise.
Materials
- Test tubes
- Micropipettes
- Sterile pipette tips
- DI water
- Hot water bath (80oC and 37 oC)
- Ice bath (4oC)
- 1% starch solution
- I2KI solution
- Well plate
- 0.5% amylase solution
- Timer
Procedures
- Label 3 test tubes #1-#3.
- Using a P-1000 micropipette, add 1.0 mL of 1% starch solution to each tube.
- Using a new tip on your P-1000 micropipette, add 3.0 mL DI water to each tube.
- Using a new tip on your P-1000 micropipette add 1.0 mL of pH 5.0 buffer to each tube.
- Place the test tubes as follows: Tube #1 in 80oC water bath, Tube #2 in 37oC water bath (or incubator), Tube #3 in crushed ice (4oC).
- Let all 3 tubes sit in the specified environments for at least 15 minutes.
- Place 2 drops of I2KI into the compartments of several rows of the test plate so that 24 compartments have clear yellow liquid in them.
- Starting with tube #1, do the following:
- Using a CLEAN NEW TIP on a P-200 micropipette, draw 50.0 microliters of liquid from the test tube and dispense it into the first compartment of the test plate containing I2KI. The mixture should turn dark blue or black. (This confirms that starch is present in your test tube before the enzyme is added to the tube.) DO NOT TOUCH THE I2KI WITH THE PIPETTE TIP! If you do, eject the tip and change to a new clean one.
- Add 400.0 µL of 0.5% amylase solution using a P-1000 micropipette. Begin recording the time in seconds the moment the amylase is added. Mix the tube contents by swirling the tube and gently tapping the bottom of the tube against your palm. Proceed to the next step immediately (within 20 seconds)
IMPORTANT! Leave the tubes in their temperature environments as they are being tested!
- At exactly 20 seconds, transfer 50.0 µL (using your P-200 micropipette) of the reaction mixture from tube #1 into the next new compartment containing I2KI on the test plate.
- Do not touch the I2KI with the Pipette Tip!
- You can continue using the same tip for step 8 below as long as it remains uncontaminated.
- Repeat step 11 every 20 seconds using the next new compartment on the test plate. Continue until a blue-black color is no longer produced and the I2KI solution remains amber (indicating that no starch remains). Then count the total number of compartments that did change colors plus on that did not change colors and multiply by 20. Record the time required for the complete digestion of the starch in Table 2.
- If the color of the I2KI continues to change to a darker color after a total of 8 minutes of testing, stop testing that reaction mixture, record 480 seconds as the time in the data table, and proceed with step 9.
- Repeat steps 9 - 13 using tube #2 then #3. You may need to clean your test plate by rinsing it with DI water, tapping it dry, and then adding fresh I2KI to the compartments.
|
Tube |
Temperature (oC) |
Time of Starch Digestion (sec) |
|---|---|---|
|
1 |
80 |
|
|
2 |
37 |
|
|
3 |
4 |
Data Analysis
How would you interpret the results shown in Table 10.2?
Study Questions
- How does an enzyme speed up a chemical reaction?
- What factors can denature a protein? How?
- What reaction does amylase catalyze?
- Explain the colormetric assay used to monitor amylase activity.


