6.4: Restriction Mapping
- Page ID
- 18157
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The restriction/modification system in bacteria is a small-scale immune system for protection from infection by foreign DNA.
In the late 1960's it was discovered that E. coli contains enzymes that will methylate specific nucleotide bases in DNA
· Different strains of E. coli contained different types of these methylases
- Typical sites of methylation include the N6 position of adenine, the N4 position of cytosine, or the C5 position of cytosine.
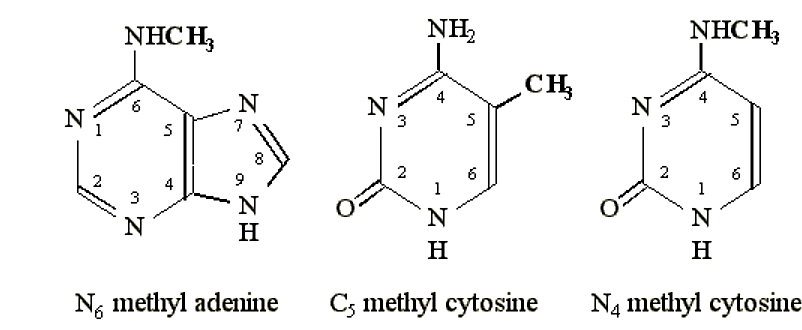.png?revision=1&size=bestfit&width=548&height=225)
Figure 6.4.1: Methylation sites
- In addition, only a fractional percentage of bases were methylated (i.e. not every adenine was methylated, for example) and these occurred at very specific sites in the DNA.
- A characteristic feature of the sites of methylation, was that they involved palindromic DNA sequences.
- Here is an example from a particular E. coli strain R1:
.png?revision=1&size=bestfit&width=450&height=115)
Figure 6.4.2: Palindromic DNA
(EcoR1 methylase specificity. Rubin and Modrich, 1977)
- In addition to possessing a particular methylase, individual bacterial strains also contained accompanying specific endonuclease activities.
- The endonucleases cleaved at or near the methylation recognition site.
Figure 6.4.3: Cleavage near methylation site
- These specific nucleases, however, would not cleave at these specific palindromic sequences if the DNA was methylated.
Thus, this combination of a specific methylase and associated endonuclease functioned as a type of immune system for individual bacterial strains, protecting them from infection by foreign DNA (e.g. viruses).
- In the bacterial strain EcoR1, the sequence GAATTC will be methylated at the internal adenine base (by the EcoR1 methylase).
- The EcoR1 endonuclease within the same bacteria will not cleave the methylated DNA.
- Foreign viral DNA, which is not methylated at the sequence "GAATTC" will therefore be recognized as "foreign" DNA and willbe cleaved by the EcoR1 endonuclease.
- Cleavage of the viral DNA renders it non-functional.
Such endonucleases are referred to as "restriction endonucleases" because they restrict the DNA within the cell to being "self".
The combination of restriction endonuclease and methylase is termed the "restriction-modification" system.
Since different bacterial strains and species have potentially different R/M systems, their characterization has made available hundreds of endonucleases with different sequence specific cleavage sites.
- They are one of the primary tools in modern molecular biology for the manipulation and identification of DNA sequences.
- Restriction endonucleases are commonly named after the bacterium from which it was isolated.
|
Name |
Source |
Recognition Sequence |
Comments |
|
Alu I |
Arthrobacter luteus |
| 5'… A G C T … 3' 3'… T C G A … 5' | |
"Four cutter". Leaves blunt ends to the DNA. |
|
Bfa I |
Bacteroides fragilis |
| 5'… C T A G … 3' 3'… G A T C … 5' | |
"Four cutter". Leaves 5' overhang. |
|
Nci I |
Neisseria cinerea |
| C 5'… C C G G G … 3' 3'… G G C C C … 5' G | |
"Five cutter". Middle base can be either cytosine or guanine. Leaves 5' overhang. Different recognition sites may have non-complementary sequences. |
|
Eco R1 |
Escherichia coli |
| 5'… G A A T T C … 3' 3'… C T T A A G … 5' | |
"Six cutter". Leaves 5' overhang. Behaves like a "four cutter" ('star' activity) in high salt buffer. $44 for 10,000 units. |
|
Hae II |
Haemophilusaegyptius |
| 5'… Pu G C G C Py … 3' 3'… Py C G C G Pu … 5' | |
"Six cutter". Pu is any purine, Py is any pyrimidine. Leaves 3' overhang. |
|
EcoO109I |
Escherichia coli |
| 5'… Pu G G N C C Py … 3' 3'… Py C C N G G Pu … 5' | |
"Seven cutter". Pu is any purine, Py is any pyrimidine, N is any base. Leaves 5' overhang. Different recognition sites may have non-complementary sequences. |
|
Bgl I |
Bacillus globigii |
| 5'… GCCN NNNNGGC … 3' 3'… CGGNNNN NCCG … 5' | |
"Six cutter with interrupted palindrome". Leaves 5' overhang. Different recognition sites may have non-complementary sequences. |
|
Bsa HI |
Bacillusstearothermophilus |
| 5'… G Pu C G Py C … 3' 3'… C Py G C Pu G … 5' | |
"Six cutter". Different recognition sites will be complementary. |
|
Aat II |
Acetobacter aceti |
| 5'… G A C G T C … 3' 3'… C T G C A G … 5' | |
"Six cutter" with 3' overhang. Same recognition sequence as Bsa HI, but different cleavage position. |
|
Bpm I |
Bacillus pumilus |
| 5'… C T G G A G N16 … 3' 3'… G A C C T C N14 … 5' | |
Non-palindrome, distal cleavage. Leaves 3' overhang. $50 for 50 units. |
|
Not I |
Nocardiaotitidiscaviarum |
| 5'… G C G G C C G C … 3' 3'… C G C C G G C G … 5' | |
"Eight cutter". Leaves 5' overhang. |
|
Bsm I |
Bacillusstearothermophilus |
| 5'… G A A T G C N … 3' 3'… C T T A C G N … 5' | |
"Weird". Leaves 3' overhang. |
- The utility of restriction endonucleases lies in their specificity and the frequency with which their recognition sites occur within any given DNA sample.
- If there is a 25% probability for a specific base at any given site, then the frequency with which different restriction endonuclease sites will occur can be easily calculated (0.25n):
|
Specificity |
Example |
Frequency of Occurrence |
|
Four base sequence |
Alu I |
1 Alu site in every 256 bases (0.25 Kb) |
|
Five base sequence |
Nci I |
1 Nci I site in every 1024 bases (1.0 Kb) |
|
Six base sequence |
EcoR I |
1 EcoR1 site in every 4,096 bases (4.1 Kb) |
|
Seven base sequence |
EcoO109I |
1 EcoO109I site in every 16,384 bases (16.4 Kb) |
|
Eight base sequence |
Not I |
1 Not I site in every 65,536 bases (65.5 Kb) |
Thus, on average, any given DNA will contain an Alu I site every 0.25 kilobases, whereas a Not I site occurs once about every 65.5 kilobases.
- Not I is therefore a very useful enzyme for isolating large regions of DNA, typically in research involving genomic DNA manipulations.
- Alu I would be expected to digest a DNA sample into lots of little pieces.
The assortment of DNA fragments would represent a specific "fingerprint" of the particular DNA being digested. Different DNA would not yield the same collection of fragment sizes. Thus, DNA from different sources can be either matched or distinguished based on the assembly of fragments after restriction endonuclease treatment. These are termed "Restriction Fragment Length Polymorphisms", or RFLP's. This simple analysis is used in various aspects of molecular biology as well as a law enforcement and genealogy. For example, genetic variations that distinguish individuals also may result in fewer or additional restriction endonuclease recognition sites.
Gel Electrophoresis of DNA
The most common gel electrophoresis solid support matrix for DNA molecules is
- agarose and
- acrylamide.
DNA agarose gels
The electrophoretic migration rate of DNA through agarose gels is dependent upon four main parameters:
1. The molecular size of the DNA. Molecules of linear duplex DNA travel through agarose gels at a rate which is inversely proportional to the log of their molecular weight.
Mr α 1/log (Mw)
Example: Compare molecular mass vs. expected migration rate:
|
Molecular Mass (Da) |
log (Molec. Mass) |
1/log (Molec. Mass) |
|
100,000 |
5.0 |
0.20 |
|
50,000 |
4.7 |
0.21 |
|
10,000 |
4.0 |
0.25 |
|
5,000 |
3.7 |
0.27 |
|
1,000 |
3.0 |
0.33 |
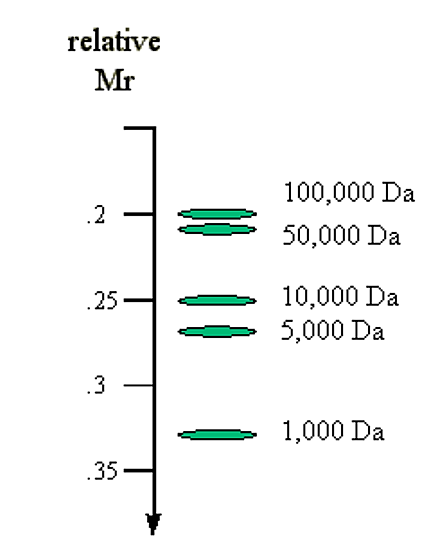.png?revision=1&size=bestfit&width=235&height=292)
Figure 6.4.4: Molecular mass and migration rate
2. The agarose concentration. There is an inverse linear relationship between the logarithm of the electrophoretic mobility and gel concentration.
|
Agarose (%) |
Range of separation of linear DNA (in kilobases) |
|
0.3 |
60 - 5 |
|
0.6 |
20 - 1 |
|
0.7 |
10 - 0.8 |
|
0.9 |
7 - 0.5 |
|
1.2 |
6 - 0.4 |
|
1.5 |
4 - 0.2 |
|
2.0 |
3 - 0.1 |
3. The conformation of the DNA.
- closed circular DNA (form-I) - typically supercoiled (compact)
- nicked circular (form-II) - nick relaxes any supercoiling
- linear DNA (form-III)
These different forms of the same DNA migrate at different rates through an agarose gel.
- Almost always the linear form (form-III) migrates at the slowest rate of the three forms
- Supercoiled DNA (form-I) usually migrates the fastest
.png?revision=1&size=bestfit&width=517&height=231)
Figure 6.4.5: Forms of DNA
4. The applied voltage.
Other details:
- Typical value for running an agarose gel is 5 volts per cm (length of gel).
- Agarose gels are usually poured and run horizontally
- Finally, the DNA being an acidic molecule, migrates towards the positively charged electrode (cathode). DNA naturally has a constant charge to mass ratio, so no detergents need to be added (as with proteins)
.png?revision=1&size=bestfit&width=576&height=417)
Figure 6.4.6: Gel electrophoresis
DNA acrylamide gels
- Acrylamide gels are useful for separation of small DNA fragments
- typically oligonucleotides <100 base pairs.
- These gels are usually of a low acrylamide concentration (<=6%) and contain the non-ionic denaturing agent Urea (6M).
- The denaturing agent prevents secondary structure formation in oligonucleotides and allows a relatively accurate determination of molecular mass.
Staining of DNA
- The most convenient method to visualize DNA in gel electrophoresis is staining with the fluorescent dye ethidium bromide.
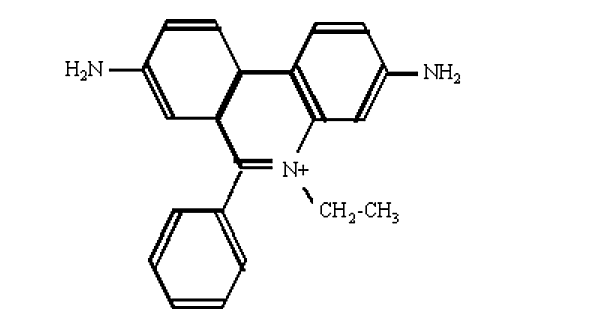.png?revision=1&size=bestfit&width=402&height=216)
Figure 6.4.7: Ethidium bromide
- This compound contains a planar group that intercalates between the stacked bases of DNA.
- The orientation and proximity of ethidium with the stacked bases causes the dye to display an increased flourescencecompared to free dye (in solution).
- U.V. radiation at 254 nm is absorbed by the DNA and transmitted to the bound dye.
- The energy is re-emitted at 590 nm in the red-orange region of the spectrum.
- Ethidium bromide is usually prepared as a stock solution of 10 mg/ml in water, stored at room temp and protected from light.
- The dye is usually incorporated into the gel and running buffer, or conversely, the gel is stained after running by soaking in a solution of ethidium bromide (0.5 ug/ml for 30 min).
- The stain is visualized by irradiating with a UV light source (i.e. using a transiluminator) and photgraphing with polaroid film.
- The usual sensitivity of detection is better than 0.1 ug of DNA.
Because ethidium is a DNA intercalating agent, it is a powerful mutagen. Incorporation of ethidium in the DNA of living organisms (i.e. you and I) can cause (unwanted) mutations.
Combining restriction endonuclease digestion with gel electrophoresis of DNA: Restriction mapping
A given sequence of DNA (e.g. a gene) will have a specific sequence, and therefore, specific restriction endonuclease sites
- The number and location of such sites is a unique and predictable property for a given DNA molecule
- The fragmentation pattern (i.e. number and size of fragments after restriction endonuclease digestion) can be characterized by gel electrophoresis as a type of "DNA fingerprint"
- Any changes in the DNA sequence of a gene can result in the elimination of particular restriction sites, and conversely, create new ones:
.png?revision=1&size=bestfit&width=660&height=257)
Figure 6.4.8: Change in restriction sites
- The gel electrophoresis fragment pattern for the human and dog albumin gene will be characteristically different from each other
- A sample of blood can potentially be identified as either human or dog by observing the restriction fragment length polymorphism
- Genetic differences between individuals can also be identified using restriction fragment length polymorphism analysis
.png?revision=1&size=bestfit&width=233&height=313)
Figure 6.4.9: Using RFLP to see genetic differences


