16: Unknown Bacteria Identification
- Page ID
- 107328
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Apply microbiological tools to isolate and identify bacterial species of unknown identities.
- Carefully document results of microbiological tests.
- Effectively collaborate with a classmate.
- Successfully identify the unknown bacterial species.
Introduction
In this project you will experience the type of process that microbiologists have traditionally used to identify a bacterial species. This will involve:
- Isolating bacteria (one species per culture - must begin with an isolated colony to insure that there is only one species) - if bacteria are not isolated, you cannot rely on the results of any of the other tests you do.
- Conducting biochemical tests to narrow down the possible species of the unknown bacteria
- Collaborating with a partner and contributing equally to the work
- Writing a scientifically-written report detailing the project, its experiments, and its results
Important
In scientific research and throughout the professional world, collaboration occurs often and is an essential skill. Employers want you to be able to collaborate effectively with others. You are building your job and career skills here
Activity Log
For this project you will work in a pair. You are BOTH responsible for contributing equally to identifying the unknown bacteria.
Keep the following activity log to show your contributions to identifying your unknown bacteria. This insures that this group project is fair and all members of the group are contributing equally to the project. You will submit your activity log in your report. If your contributions to the project don't demonstrate equal contributions, deductions will be made to the report.
|
Date |
Task |
Student(s) In the Group Working On This Task |
Instructor Signature |
|---|---|---|---|
|
|
|||
|
|
|||
|
|
|||
|
|
|
||
|
|
|||
|
|
|||
|
|
|||
|
|
|||
|
|
|||
|
|
|||
|
|
|||
|
|
|||
|
|
|||
|
|
|
Instructions for Identifying Unknown Bacteria
- Obtain a culture of unknown bacteria from your instructor. Your unknown culture will have a letter or a number. What letter or number is your unknown bacterial culture?
2. Your culture of unknown bacteria will have two different species in it. Follow the steps below carefully to identify these bacterial species.
Step 1: Isolate the Two Bacterial Species & Do Initial Gram Stain
- Conduct four (4) streak plates on TSA in order to obtain isolated colonies (each partner will conduct two). By doing four streak plates, this increases the chances that you and your partner will obtain isolated colonies of both your bacterial species.
- Make sure your names, unknown number/letter, and the date are on the Petri plates, invert the Petri plates, tape together, and put into the bin to be incubated.
- Do a Gram stain to determine:
- Whether the mixture of bacterial species contains a Gram positive and a Gram negative species, both Gram positive, or both Gram negative.
- The cell shape of each bacterial species (i.e. coccus, bacillus, vibrio, spirillum, or spirochete).
- The cell arrangement of each bacteria species (i.e. single, pairs, chains (strepto-), or bunches (staphylo-).
- Record results in the table below. Refer back to this table in Step 2 to make sure that the bacteria you isolate match the bacteria you saw in this Gram stain.
|
Unknown Bacterial Species #1 |
Unknown Bacterial Species #2 |
|
|---|---|---|
|
Gram |
||
|
cell shape |
||
|
cell arrangement |
Note
Taking photos of results at every stage of the project will make a stand-out report! Include these photos in your report to provide visuals and evidence of your results. Make sure each photo is accompanied by a caption telling what the photo shows. If the photo is of a microscopic sample, make sure to indicate the magnification used when the photo was taken.
Step 2: Characterize Bacterial Colonies, Create Stock Cultures of Isolated Bacteria, & Do Gram Stain on Isolates
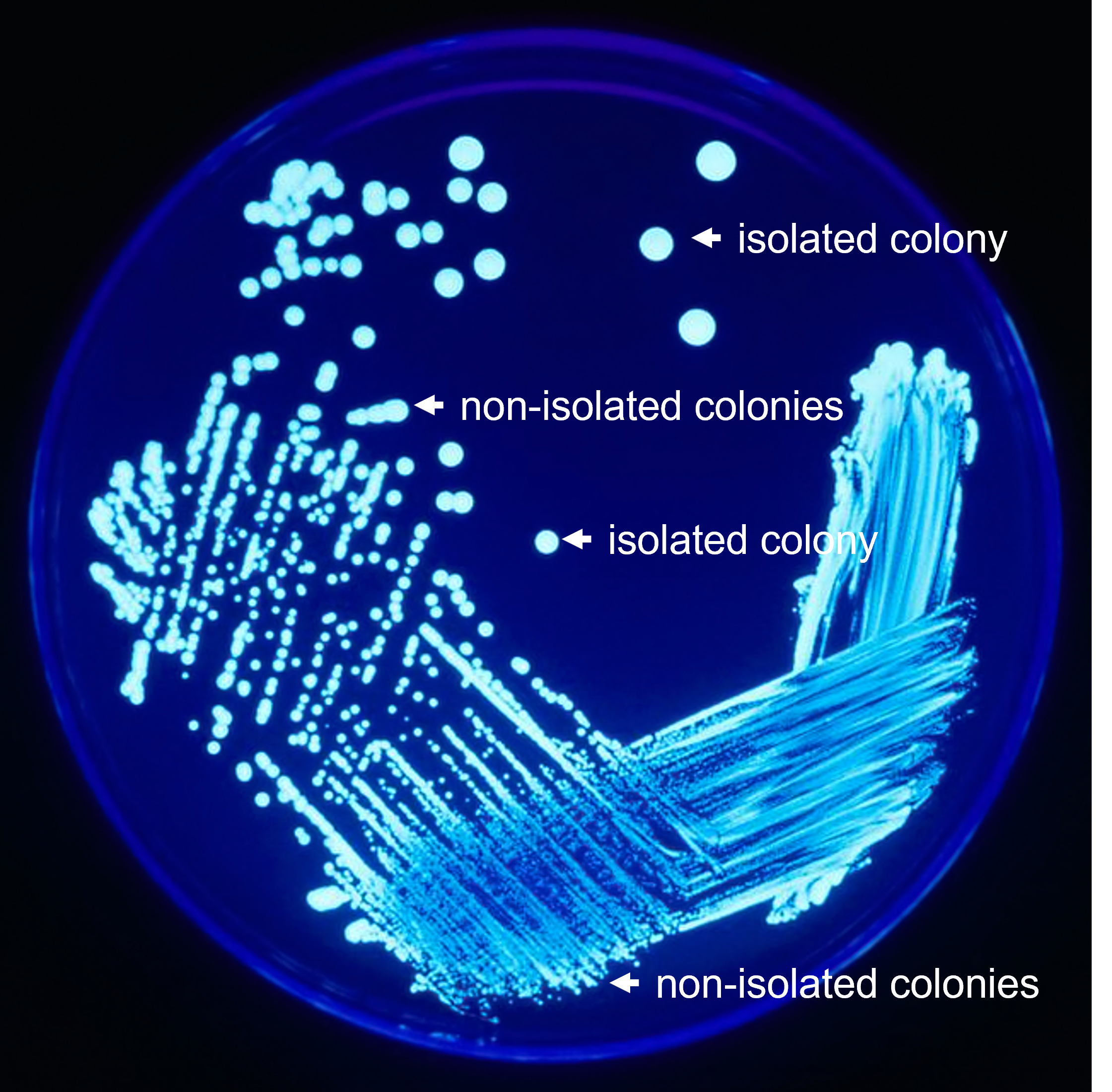 Figure 1: A quadrant streak plate after growth. Use this as a guide to help identify what are and are not isolated colonies. Isolated colonies originate from a single cell, and therefore should contain only one bacterial species. Therefore, growing a culture from an isolated colony is an effective way to separate different species.
Figure 1: A quadrant streak plate after growth. Use this as a guide to help identify what are and are not isolated colonies. Isolated colonies originate from a single cell, and therefore should contain only one bacterial species. Therefore, growing a culture from an isolated colony is an effective way to separate different species.Note
Taking photos of results at every stage of the project will make a stand-out report! Include these photos in your report to provide visuals and evidence of your results. Make sure each photo is accompanied by a caption telling what the photo shows. If the photo is of a microscopic sample, make sure to indicate the magnification used when the photo was taken.
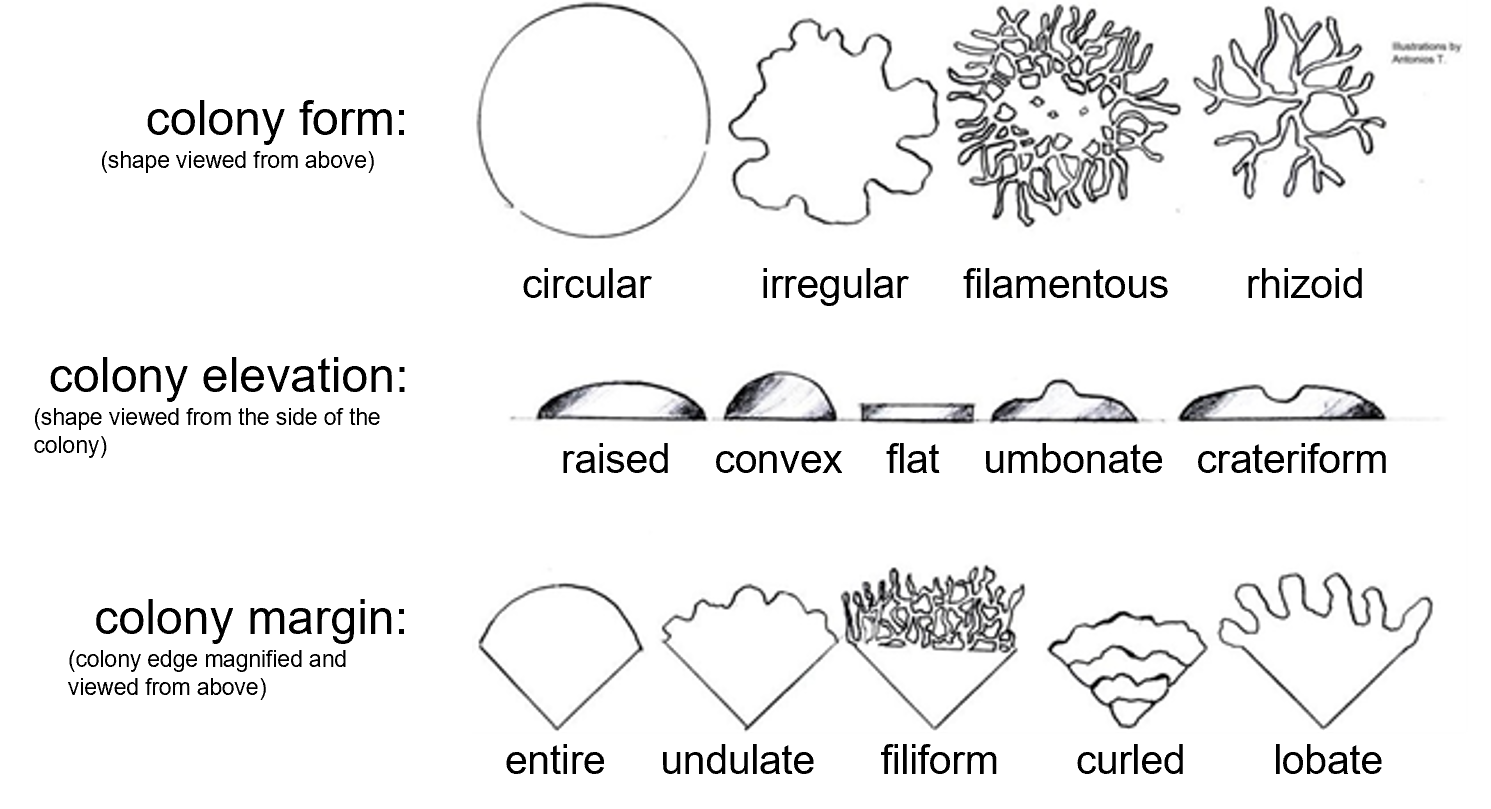
Figure 2: A guide for classifying colony characteristics. Colony form is the overall shape of the colony (circular, irregular, filamentous, or rhizoid) when viewed from above. Colony elevation examines the height and shape of the growth above the surface of the petri plate agar (raised, convex, flat, umbonate, or crateriform) and is best determined by viewing the colony from its side. Colony margin examines the shape of the edge of the colony when magnified (entire, undulate, filiform, curled, or lobate).
- Carefully examine streak plates. Identify two bacterial colonies that have differences in appearance.
- For the two different bacterial colonies you find, describe differences in the colony forms in the tables below. Do the best you can keeping your Petri plate closed to prevent contamination. It is essential that these bacterial colonies do not become contaminated with other bacteria or microbes floating in the air.
- Label two TSA slants. Give each Unknown bacterial species a name (you can use names "A" and "B" or "1" and "2" or "Yessica" and "Yoli" - choose any names you and your partner would like - you will just need to keep straight which colony is which).
- Transfer one third (1/3) of one of the isolated colonies to one of the TSA slants.
- Transfer one third (1/3) of the other isolated colony to one of the TSA slants.
- Use one third (1/3) of each of the colonies to make separate bacterial smears on a slide and conduct a Gram stain.
- Compare results from the Gram stains. Did you obtain the same types of bacteria and separate them successfully? If you see Gram positive and Gram negative cells together, they are not isolated. If you see bacilli and cocci together, they are not isolated.
- If one or both bacterial species are not isolated (they are mixed with another species), re-examine the petri plates to see if there is another colony that appears different and create a new TSA slant, and create a new bacterial smear on a slide. Do the Gram stain on this if there is still time. If there is not time, save your bacterial smear for next class to see if the bacteria are now isolated.
- If you do not have better isolated colonies or continue to find that you have a mixture, conduct more streak plates to isolate bacterial colonies again.
- When you have successfully isolated your bacteria, record results from the Gram stain in the tables below.
Step 3: Follow the Flow Chart to Identify Bacterial Species
Note
Taking photos of results at every stage of the project will make a stand-out report! Include these photos in your report to provide visuals and evidence of your results. Make sure each photo is accompanied by a caption telling what the photo shows. If the photo is of a microscopic sample, make sure to indicate the magnification used when the photo was taken.
|
Unknown Bacterial Species ___________________ (put name you chose in the blank space above) |
|
|---|---|
|
Gram |
|
|
cell shape |
|
|
cell arrangement |
|
|
colony form |
|
|
colony elevation |
|
|
colony margin |
|
|
colony color |
|
|
colony diameter (mm) |
|
|
test 1: |
|
|
test 2: |
|
|
test 3: |
|
|
bacterial species |
|
Unknown Bacterial Species ___________________ (put name you chose in the blank space above) |
|
|---|---|
|
Gram |
|
|
cell shape |
|
|
cell arrangement |
|
|
colony form |
|
|
colony elevation |
|
|
colony margin |
|
|
colony color |
|
|
colony diameter (mm) |
|
|
test 1: |
|
|
test 2: |
|
|
test 3: |
|
|
bacterial species |
- After your bacteria have been isolated and you have good results from your Gram stain, begin to follow the flow chart below.
- For example, if your Bacteria "A" is Gram-positive, use your TSA stock slant to inoculate a starch plate to do a starch hydrolysis test.
- For example, if your Bacteria "B" is Gram-negative, use your TSA stock slant to to inoculate a SIM deep culture to test for H2S production.
- Use the results (positive or negative) from each test to determine the next test you should do on the flow chart.
- Return to the appropriate chapters in this lab manual to assist with conducting each test:

Figure 3: Unknown bacterial species identification flow chart. Once your bacterial species are isolated and you have good Gram stain results, begin to follow the flow chart for both of your unknown bacterial species. The tests that are a part of this project are in boxes. Use the results from each test to determine what the next test is or what your unknown bacterial species is.
Attributions
- This page titled 7: DNA, RNA, and DNA Replication is shared under a CC BY-NC-SA 4.0 license and was authored, remixed, and/or curated by Rosanna Hartline.
- Laboratory Exercises in Microbiology: Discovering the Unseen World Through Hands-On Investigation by Joan Petersen and Susan McLaughlin is licensed under CC BY-NC-SA 4.0
- Legionella Plate 01.png by CDC/James Gathany is in the public domain.

