1.19: Cytochrome c Oxidase
- Page ID
- 79450
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Describe the role of cytochrome c oxidase in bacterial cells.
- Tell that cytochrome c oxidase is found in only certain species of bacteria and is therefore useful for bacterial species identification and characterization.
- Explain what being "oxidase positive" means about a bacterial species metabolism, electron transport chain, and genetics.
- Explain that being "oxidase negative" does not mean the species is anaerobic, but just that it does not contain the cytochrome c oxidase enzyme and gene.
- Successfully conduct and interpret an oxidase test.
Cytochrome c Oxidase
Aerobic respiration is an O2-requiring process that uses energy from nutrient molecules to produce ATP molecules to provide for the cell's energy needs. During aerobic respiration, the electron transport chain transfers high-energy electrons from protein to protein and uses that energy to build up a H+ gradient that is utilized by ATP synthase to make ATP. At the end of the aerobic electron transport chain, an enzyme transfers the electron from the electron transport chain to O2 (the electrons at the end of the chain are low energy). In some bacteria capable of aerobic respiration, that enzyme that transfers electrons to O2, the final electron acceptor for aerobic respiration, is called cytochrome c oxidase.
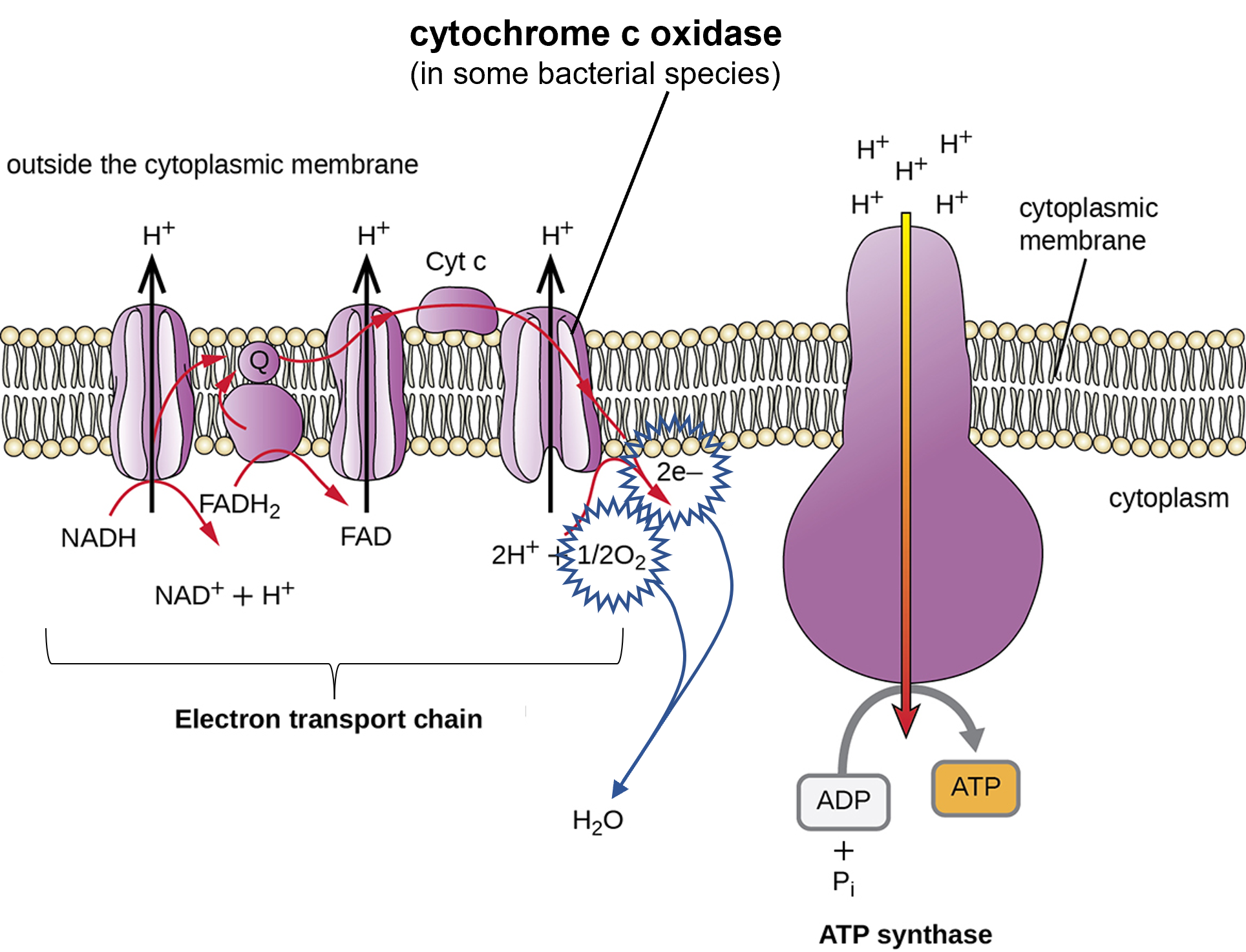
Figure 1: The bacterial electron transport chain is a series of protein complexes, electron carriers, and ion pumps that are used to pump H+ out of the bacterial cytoplasm into the extracellular space. The last enzyme in the electron transport chain can be (depends on the species) cytochrome c oxidase. The role cytochrome c oxidase plays is to remove electrons from the chain by transferring them to O2 with H+ to produce water. In aerobic bacterial species that do not have cytochrome c oxidase, this process still occurs, but using an enzyme that has a different structure and different name than cytochrome c oxidase. The gradient of H+ produced by the electron transport chain is used by the ATP synthase to make ATP. H+ flows back down the electrochemical gradient into the bacterial cytoplasm through ATP synthase, providing the energy for ATP production by oxidative phosphorylation.(credit: modification of work by Klaus Hoffmeier)
A test called the oxidase test can be used in the laboratory to determine if a bacterial species has cytochrome c oxidase. Those species that have cytochrome c oxidase are called oxidase positive and those species that do not have cytochrome c oxidase are called oxidase negative. The ability of a bacterial species to produce cytochrome c oxidase is coded in its DNA. If the cytochrome c oxidase gene is present in the bacterial species, it will produce this enzyme. If the gene is absent, that bacterial species cannot produce cytochrome c oxidase. The oxidase test is therefore useful for characterizing bacterial species and differentiating bacterial species from each other for identification purposes.
Bacterial species that contain cytochrome c oxidase are capable of aerobic respiration. Oxidase positive bacterial species are all aerobic.
However, oxidase negative species are not necessarily anaerobic. Oxidase negative species may still be aerobic species or facultative anaerobes, but they produce and use a different enzyme, other than cytochrome c oxidase, to transfer electrons from the electron transport chain to O2.
The Oxidase Test
The oxidase test is a key test to differentiate between the bacterial families Pseudomonadaceae (oxidase positive species) and Enterobacteriaceae (oxidase negative species), and is useful for speciation and identification of many other bacteria.
The oxidase test utilizes a special reagent called oxidase reagent. This reagent is colorless. However, in the presence of cytochrome c oxidase, the oxidase reagent will transfer its electrons to cytochrome c oxidase and exhibit a bluish or purplish color in 30 seconds or less.
- oxidase positive bacteria contain cytochrome c oxidase and produce a change in color of the reagent from colorless to bluish or purplish in less than 30 seconds.
- oxidase negative bacteria do not contain cytochrome c oxidase and do not change the color of oxidase reagent in less than 30 seconds.

Figure 2: Results from the oxidase test. (Left) An oxidase test strip containing oxidase reagent is still white indicating bacteria is oxidase negative. (Right) The oxidase test strip containing oxidase reagent is purple indicating the bacteria are oxidase positive. The test only indicates oxidase positive if the purple color appears in 30 seconds or less.
Laboratory Instructions
Oxidase Test
- Obtain an oxidase test strip and half of an empty petri plate.
- Place the oxidase test strips face up on the half empty petri plate.
- Lightly moisten the oxidase test strip with a small drop of water. Do not over-moisten the strip!
- Using a sterile swab or sterile loop, obtain a large amount of bacteria from a petri plate.
- Rub the bacteria on the swab or sterile loop onto the moistened region of the oxidase test strip.
- Start a timer for 30 seconds.
- Watch for purple color to develop on the oxidase test strip within 30 seconds.
- purple or bluish color change in less than 30 seconds is oxidase positive
- no color change within 30 seconds is oxidase negative (color change after 30 seconds is considered oxidase negative)
- Repeat for each bacterial species you are testing.
Results & Questions
|
|
Oxidase (+/-) |
Species produces cytochrome c oxidase (+/-) |
|---|---|---|
|
Escherichia coli |
|
|
|
Pseudomonas aeruginosa |
|
- Complete the table above with results of the oxidase test.
- What metabolic pathway is cytochrome c oxidase important in?
- What would happen to a bacterial cell that lost its ability to produce cytochrome c oxidase?
- Is a bacterial species that produces cytochrome c oxidase aerobic? Explain your answer.
- Is a bacterial species that does not produce cytochrome c oxidase anaerobic? Explain your answer.
- Is the oxidase test useful for bacterial species identification and characterization? Explain your answer.
Attributions
- Chapter Image: Red Mountain Microbiology by Jill Raymond Ph.D.; Graham Boorse, Ph.D.; and Anne Mason M.S. is licensed under CC BY-NC 4.0
- MB352 General Microbiology Laboratory 2021 (Lee) by Alice_Lee@ncsu.edu is licensed under CC BY-NC-SA 4.0
- Microbiology by OpenStax is licensed under CC BY 4.0
- Red Mountain Microbiology by Jill Raymond Ph.D.; Graham Boorse, Ph.D.; Anne Mason M.S. is licensed under CC BY-NC 4.0


