10: DNA, RNA, and DNA Replication
- Page ID
- 106459
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Describe and identify the structures of DNA from nucleotide components to double helix.
- Describe and identify the structures of RNA from nucleotide components to single strand.
- Compare the structures of DNA and RNA.
- Utilize and identify 5' and 3' directionality of DNA and RNA nucleotides and molecules.
- Explain the process of DNA replication in detail and identify components of the process in diagrams and figures.
- Build DNA and RNA nucleotides using puzzle pieces and compare their structures.
- Build DNA and RNA nucleic acid sequences using puzzle pieces and identify and compare structures of these molecules.
- Replicate the process of DNA replication using a DNA puzzle model.
- Compare the puzzle re-creation of DNA replication with events that occur in the actual process of DNA replication.
Importance of Understanding DNA Structure
Understanding the structure of DNA is fundamental to a better understanding of biology overall, but particularly microbiology, disease, and modern medical approaches. Here are a few ways that having a thorough understanding of DNA structure will impact your ability to understand additional microbiological topics:
- the DNA of a microbe dictates if that microbe is pathogenic or not and the degree of its pathogenicity
- the DNA of a microbe is the basic blueprint for that microbe's metabolism, characteristics, abilities, structure, and survival approaches
- DNA information is used during the process of gene expression
- understanding DNA structure is essential to better understand gene expression
- understanding how gene expression works enables us to understand the link between DNA and a microbe's characteristics
- how pathogens (disease-causing microbes) evolve/change over time (think of the seasonal flu strains or new variants of the virus that causes COVID-19) is related to its DNA (or the closely related molecule RNA in the case of the virus that causes COVID-19) and how that DNA changes over time
- DNA is used to identify microbes (e.g. what microbe is causing an infection) in diagnostic approaches such as PCR testing
- understanding how PCR works for diagnostic techniques requires a firm understanding of DNA structure and DNA replication
- understanding modern biological science is impossible without a firm understanding of DNA structure and DNA replication (and PCR too)
PCR has a huge number of applications beyond medical diagnostics!
DNA Structure
Nucleotides are the Monomers of DNA
The building blocks of DNA are nucleotides. The important components of the nucleotide are a nitrogenous (nitrogen-bearing) base, a 5-carbon sugar (pentose), and a phosphate group. The nucleotide is named depending on the nitrogenous base. The nitrogenous base can be a purine such as adenine (A) and guanine (G), or a pyrimidine such as cytosine (C) and thymine (T).
The purines have a double ring structure with a six-membered ring fused to a five-membered ring. Pyrimidines are smaller in size; they have a single six-membered ring structure.
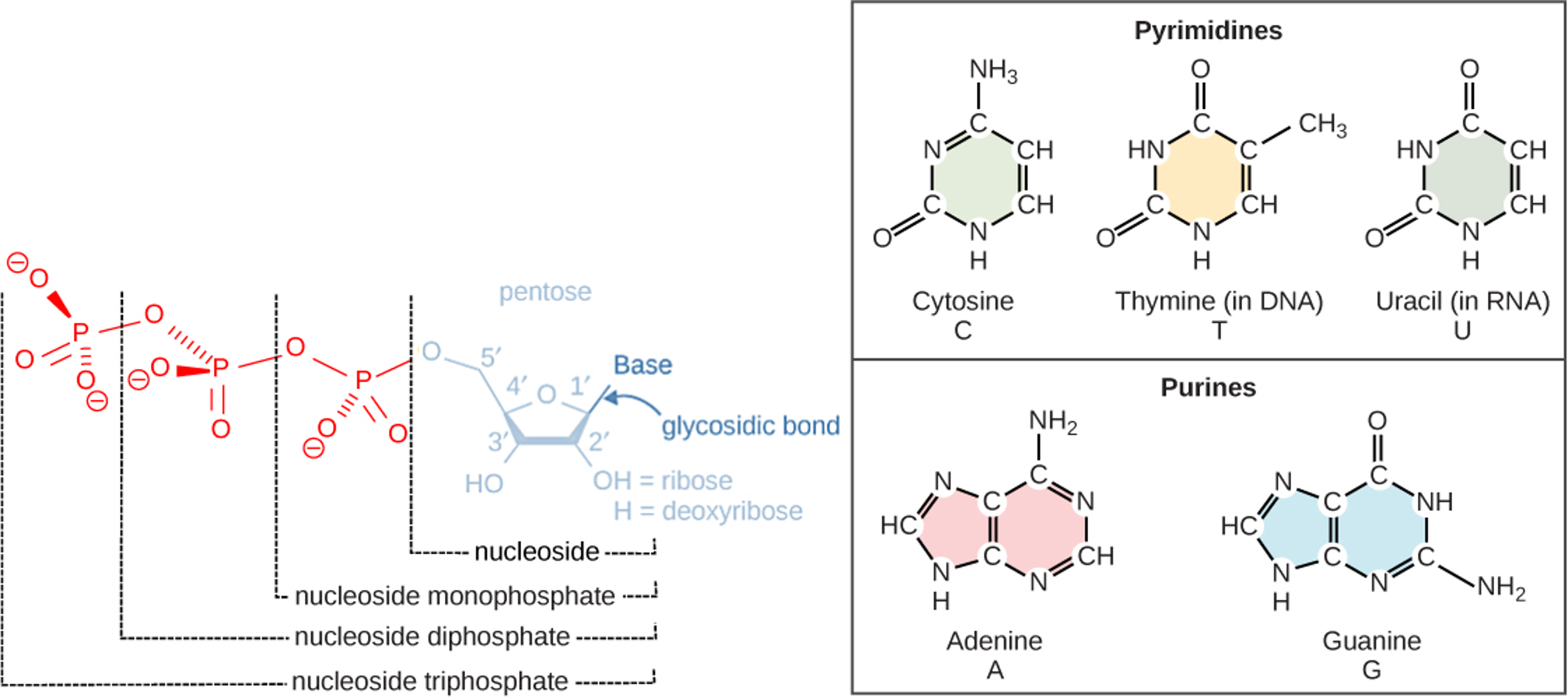
Figure 1: Nucleotides are the building blocks of DNA and RNA. This figure shows the generalized structure of nucleotides and their bases. (Left) A single nucleotide is made of a phosphate group, a pentose sugar (a sugar with five carbons) and a base. The pentose sugar is deoxyribose in DNA and has a hydrogen attached to the 2' carbon atom instead of an OH. The pentose sugar is ribose in RNA and has an OH attached to the 2' carbon atom instead of just a hydrogen. The bases found in the nucleotides can be either classified as purines or pyrimidines. (Right) The purines have a double ring structure with a six-membered ring fused to a five-membered ring. Pyrimidines are smaller in size; they have a single six-membered ring structure. Each base contains at least two nitrogen atoms. It is because of this that the bases are commonly referred to as "nitrogenous bases."
The carbon atoms of the pentose sugar are numbered 1', 2', 3', 4', and 5' (1' is read as “one prime,” 2' is read "two prime," etc.). These prime numbers are used to describe the direction of DNA strands using 5' to designate one side of the DNA molecule and using 3' to designate the other side of the DNA molecule.
When nucleotides are built in processes such as DNA replication and gene expression, these directions are incredibly important since nucleic acids can only form new bonds on the 3' end. As a result, it is often stated that nucleic acids (DNA and RNA) are built from 5' to 3'.
The sugar is deoxyribose in DNA and ribose in RNA. The phosphate, which makes DNA and RNA acidic, is connected to the 5' carbon of the sugar by the formation of an ester linkage between phosphoric acid and the 5'-OH group (an ester is an acid + an alcohol). In DNA nucleotides, the 3' carbon of the sugar deoxyribose is attached to a hydroxyl (OH) group. In RNA nucleotides, the 2' carbon of the sugar ribose also contains a hydroxyl group. The base is attached to the 1' carbon of the sugar.
Overall DNA Structure
The nucleotides form covalent bonds with each other to produce phosphodiester bonds (a fancy science name for the covalent bonds joining nucleotides together). The phosphate group forms a covalent bond with the hydroxyl group of the 3' carbon of the sugar of the next nucleotide, thereby forming a 5'-3' phosphodiester bond. In a polynucleotide, one end of the chain has a free 5' phosphate, and the other end has a free 3'-OH. These are called the 5' and 3' ends of the chain.
The result of nucleotides bonding together is a sugar-phosphate backbone with the nitrogenous bases hanging off of the side. This structure resembles one half of a ladder. The side support rails of the ladder is analagous to the sugar-phosphate backbone, and the half-rung in the middle of the ladder is analagous to the bases hanging off the side. In order for DNA to be a complete double-stranded molecule, the single DNA strand pairs with another DNA strand. The result is a complete ladder with the side-supports rails being sugar-phosphate backbones and the rungs in the middle are the bases of the two DNA strands interacting with each other in the middle.
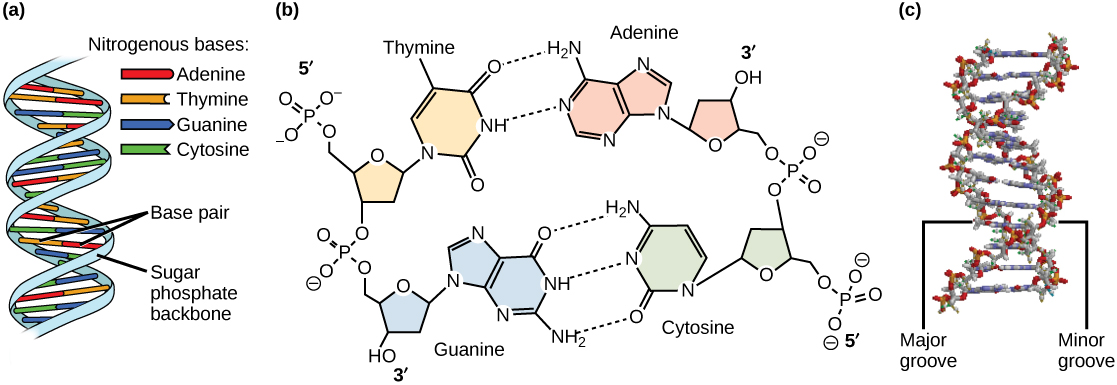
Figure 2: DNA has (a) a double helix structure and (b) phosphodiester bonds; the dotted lines between thymine and adenine and guanine and cytosine represent hydrogen bonds. The (c) major and minor grooves are binding sites for DNA binding proteins during processes such as transcription (the copying of RNA from DNA) and replication.
The two DNA strands can only pair together when the two strands are antiparallel, that is, the strands are parallel, but in opposite directions. One DNA strand will be in the 5' to 3' direction, and it will pair with a strand upside-down to it in the 3' to 5' position. This arrangement enables complementary base pairs to hydrogen bond with each other. The DNA bases that meet in the middle of the double-stranded DNA molecule pair such that adenine (A) always pairs with thymine (T) and guanine (G) always pairs with cytosine (C). The A-T pair is held together by two hydrogen bonds and the G-C pair is held together by three hydrogen bonds.
The entire DNA ladder twists in three-dimensions to produce a helical formation to the molecule.
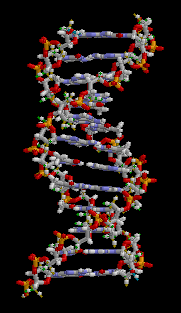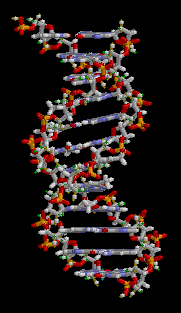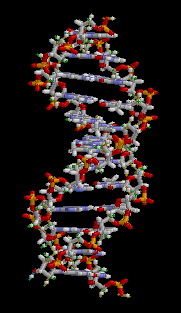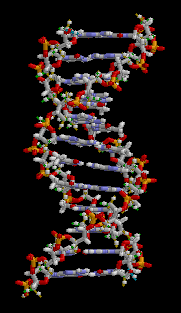
Figure 3: This animation shows the three-dimensional structure of a DNA double helix. Notice how this molecule appears similar to a ladder that has been twisted. The rungs of the ladder are the A-T and G-C bases of the two DNA strands forming hydrogen bonds with each other. The supportive side rails of the ladder are the sugar-phosphate backbones of the two DNA strands.
RNA Structure
DNA structure is very similar to DNA structure. RNA is composed of nucleotides bonded together with phosphodiester bonds forming a sugar-phosphate backbone with bases hanging off of the side. There are three main differences between DNA and RNA:
- RNA nucleotides contain ribose as the pentose sugar instead of deoxyribose (ribose has an -OH on the 2' carbon whereas deoxyribose has an -H on the 2' carbon)
- RNA nucleotides will not contain the base thymine, but will instead contain uracil; uracil base-pairs with adenine in the same way thymine does
- RNA is a single-stranded molecule
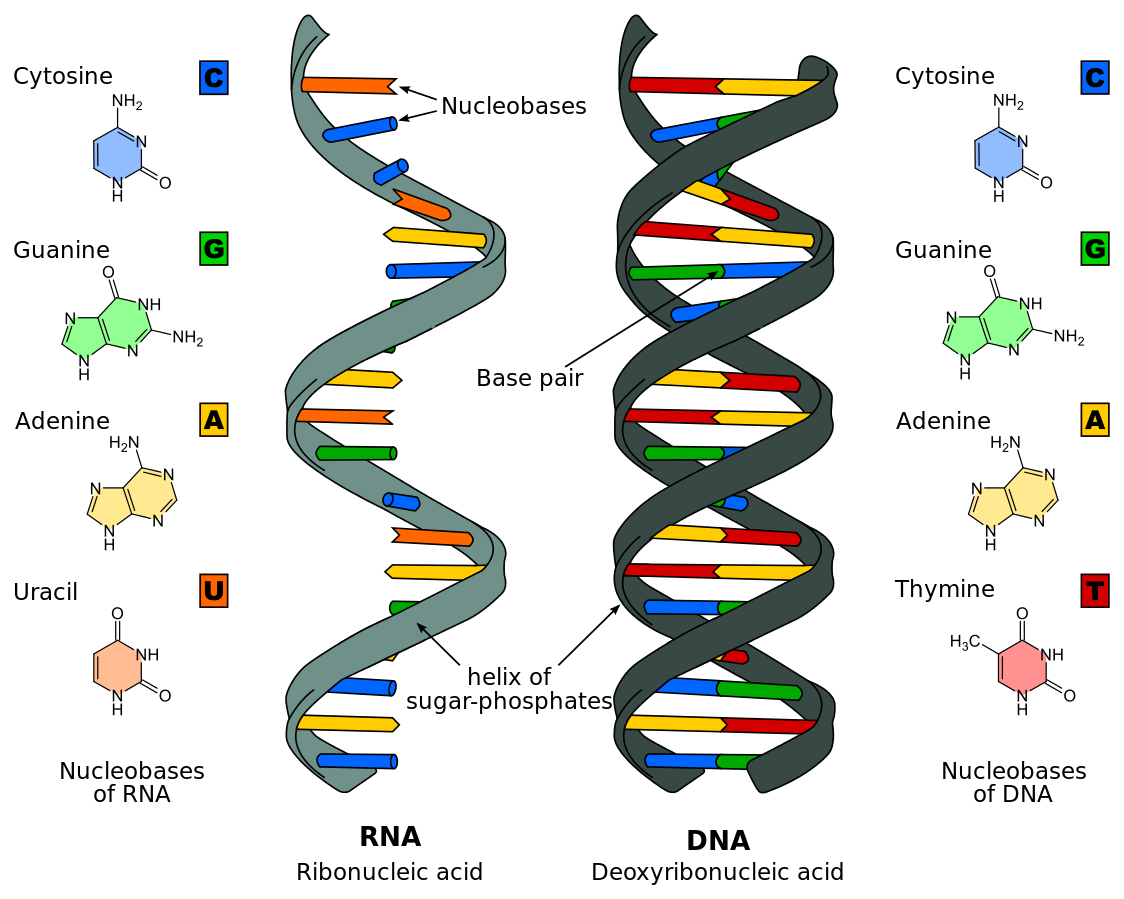
Figure 4: A comparison between the structure of DNA and the structure of RNA. This diagram shows how RNA is single-stranded and DNA is double-stranded, and that RNA contains uracil instead of thymine.
DNA Replication
DNA replication has been well studied in prokaryotes primarily because of the small size of the genome and because of the large variety of mutants that are available. E. coli has 4.6 million base pairs in a single circular chromosome and all of it gets replicated in approximately 42 minutes, starting from a single site along the chromosome and proceeding around the circle in both directions. This means that approximately 1000 nucleotides are added per second. Thus, the process is quite rapid and occurs without many mistakes.
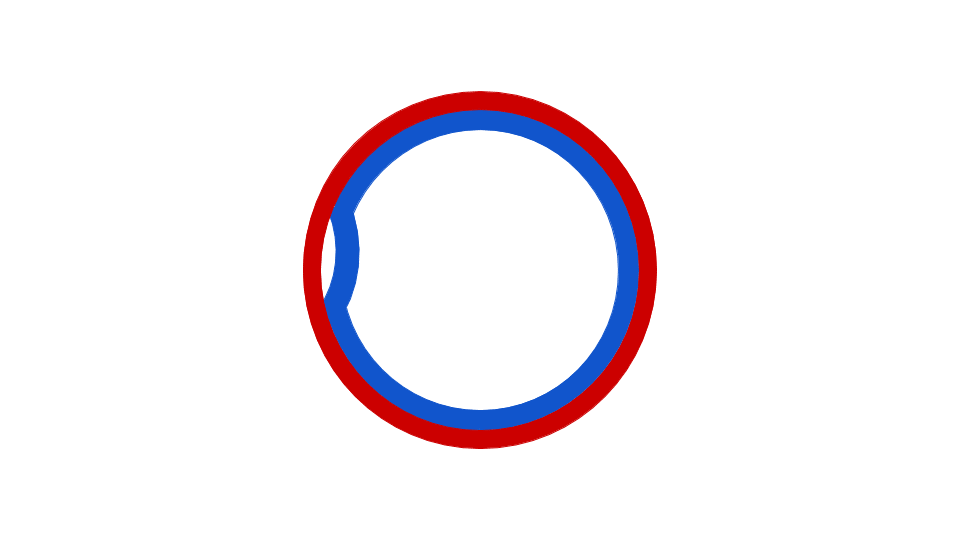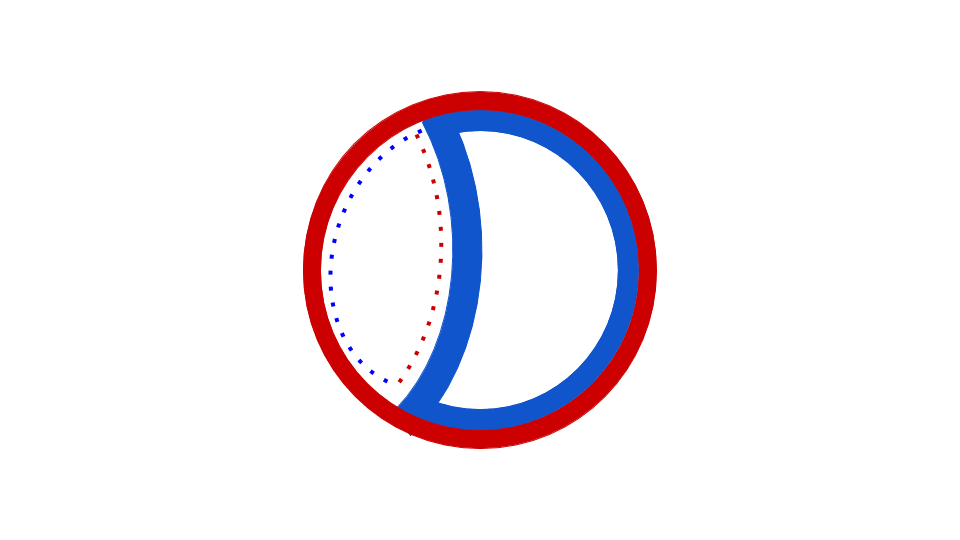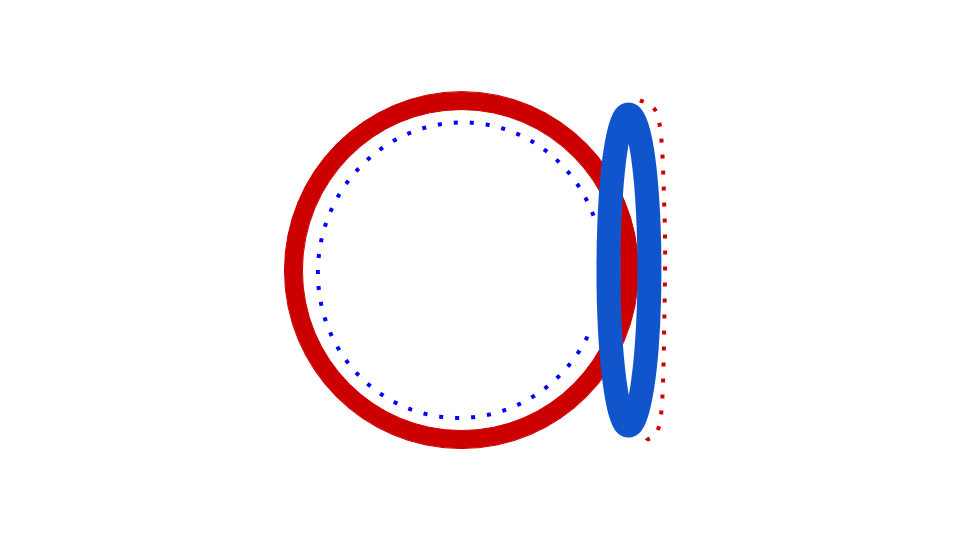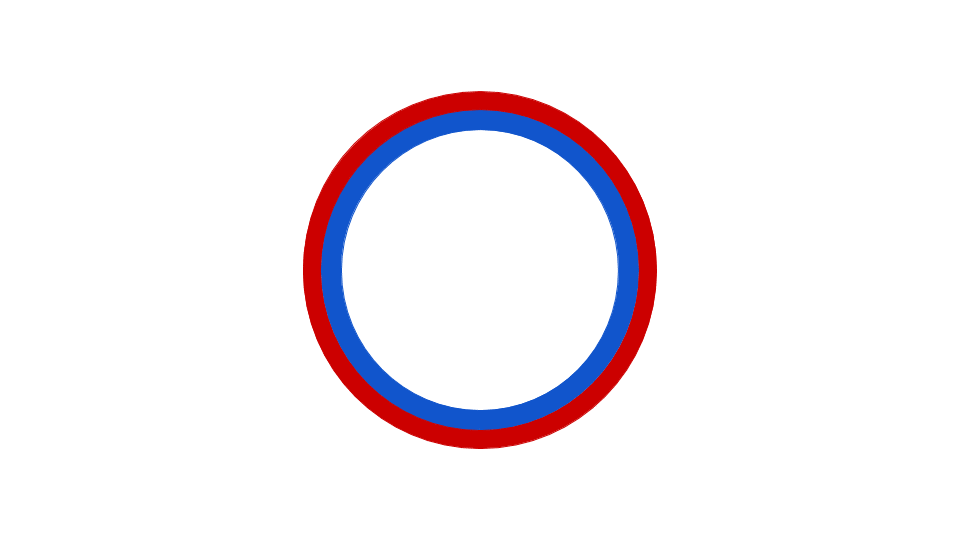
Figure 5: This animation shows how the circular bacterial chromosome is duplicated during DNA replication. The solid red circle and solid blue circle represent the two original DNA strands that are paired together in the bacterial chromosome. DNA replication begins at the origin of replication. DNA is gradually built to complement both the original DNA strands simultaneously (dotted red and dotted blue represent the new DNA being built to complement the original DNA strands). The end result is two bacterial chromosomes, each with one "old" and one "new" DNA strand. This is why the process of DNA replication is called semiconservative. One old strand is retained and paired with a new strand in each DNA molecule.
DNA replication employs a large number of structural proteins and enzymes, each of which plays a critical role during the process. One of the key players is the enzyme DNA polymerase, also known as DNA pol, which adds nucleotides one-by-one to the growing DNA chain that is complementary to the template strand. The addition of nucleotides requires energy; this energy is obtained from the nucleoside triphosphates ATP, GTP, TTP and CTP. Like ATP, the other NTPs (nucleoside triphosphates) are high-energy molecules that can serve both as the source of DNA nucleotides and the source of energy to drive the polymerization. When the bond between the phosphates is “broken,” the energy released is used to form the phosphodiester bond between the incoming nucleotide and the growing chain. In prokaryotes, three main types of polymerases are known: DNA pol I, DNA pol II, and DNA pol III. It is now known that DNA pol III is the enzyme required for DNA synthesis; DNA pol I is an important accessory enzyme in DNA replication, and along with DNA pol II, is primarily required for repair.
How does the replication machinery know where to begin? It turns out that there are specific nucleotide sequences called origins of replication where replication begins. In E. coli, which has a single origin of replication on its one chromosome (as do most prokaryotes), this origin of replication is approximately 245 base pairs long and is rich in AT sequences. The origin of replication is recognized by certain proteins that bind to this site. An enzyme called helicase unwinds the DNA by breaking the hydrogen bonds between the nitrogenous base pairs. ATP hydrolysis is required for this process. As the DNA opens up, Y-shaped structures called replication forks are formed. Two replication forks are formed at the origin of replication and these get extended bi-directionally as replication proceeds. Single-strand binding proteins coat the single strands of DNA near the replication fork to prevent the single-stranded DNA from winding back into a double helix.
DNA polymerase has two important restrictions: it is able to add nucleotides only in the 5' to 3' direction (a new DNA strand can be only extended in this direction). It also requires a free 3'-OH group to which it can add nucleotides by forming a phosphodiester bond between the 3'-OH end and the 5' phosphate of the next nucleotide. This essentially means that it cannot add nucleotides if a free 3'-OH group is not available. Then how does it add the first nucleotide? The problem is solved with the help of a primer that provides the free 3'-OH end. Another enzyme, RNA primase, synthesizes an RNA segment that is about five to ten nucleotides long and complementary to the template DNA. Because this sequence primes the DNA synthesis, it is appropriately called the primer. DNA polymerase can now extend this RNA primer, adding nucleotides one-by-one that are complementary to the template strand.
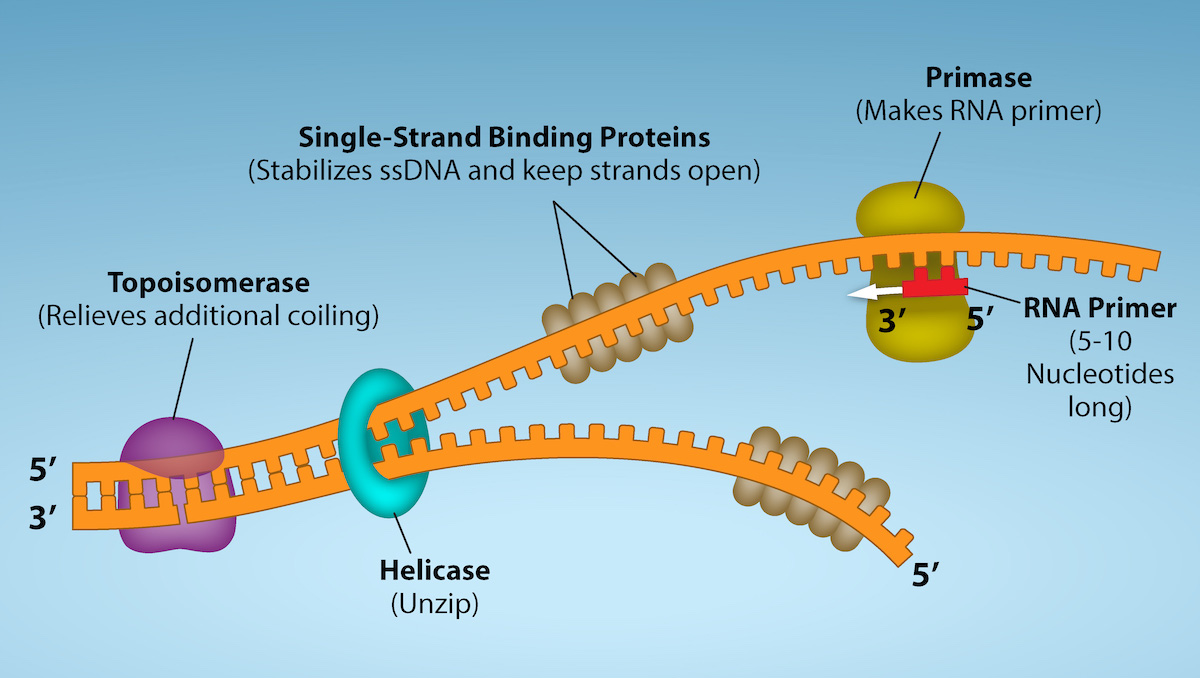
Figure 6: First Components of DNA Replication. As DNA replication begins, DNA Helicase, a large enzyme, separates the two strands of DNA so that they can act as templates for replication. Single-strand binding proteins bind to each strand to stabilize and prevent them from reforming the double helix. Primase, an RNA polymerase, binds to the single stranded DNA and synthesizes a short RNA primer in the 5’ to 3’ direction that is antiparallel to the parental strand. This RNA primer allows for DNA polymerase to begin replicating the DNA. Topoisomerase binds to the double helix upstream of the replication fork to prevent additional coiling by making small cuts in one of the DNA strands. Credit: Rao, A., Ryan, K. Fletcher, S. and Tag, A. Department of Biology, Texas A&M University.
The replication fork moves at the rate of 1000 nucleotides per second. Topoisomerase prevents the over-winding of the DNA double helix ahead of the replication fork as the DNA is opening up; it does so by causing temporary nicks in the DNA helix and then resealing it. Because DNA polymerase can only extend in the 5' to 3' direction, and because the DNA double helix is antiparallel, there is a slight problem at the replication fork. The two template DNA strands have opposing orientations: one strand is in the 5' to 3' direction and the other is oriented in the 3' to 5' direction. Only one new DNA strand, the one that is complementary to the 3' to 5' parental DNA strand, can be synthesized continuously towards the replication fork. This continuously synthesized strand is known as the leading strand. The other strand, complementary to the 5' to 3' parental DNA, is extended away from the replication fork, in small fragments known as Okazaki fragments, each requiring a primer to start the synthesis. New primer segments are laid down in the direction of the replication fork, but each pointing away from it. (Okazaki fragments are named after the Japanese scientist who first discovered them. The strand with the Okazaki fragments is known as the lagging strand.)
The leading strand can be extended from a single primer, whereas the lagging strand needs a new primer for each of the short Okazaki fragments. The overall direction of the lagging strand will be 3' to 5', and that of the leading strand 5' to 3'. A protein called the sliding clamp holds the DNA polymerase in place as it continues to add nucleotides. The sliding clamp is a ring-shaped protein that binds to the DNA and holds the polymerase in place. As synthesis proceeds, the RNA primers are replaced by DNA. The primers are removed by the exonuclease activity of DNA pol I, which uses DNA behind the RNA as its own primer and fills in the gaps left by removal of the RNA nucleotides by the addition of DNA nucleotides. The nicks that remain between the newly synthesized DNA (that replaced the RNA primer) and the previously synthesized DNA are sealed by the enzyme DNA ligase, which catalyzes the formation of phosphodiester linkages between the 3'-OH end of one nucleotide and the 5' phosphate end of the other fragment.
Once the chromosome has been completely replicated, the two DNA copies move into two different cells during cell division.
The process of DNA replication can be summarized as follows:
- DNA unwinds at the origin of replication.
- Helicase opens up the DNA-forming replication forks; these are extended bidirectionally.
- Single-strand binding proteins coat the DNA around the replication fork to prevent rewinding of the DNA.
- Topoisomerase binds at the region ahead of the replication fork to prevent supercoiling.
- Primase synthesizes RNA primers complementary to the DNA strand.
- DNA polymerase III starts adding nucleotides to the 3'-OH end of the primer.
- Elongation of both the lagging and the leading strand continues.
- RNA primers are removed by exonuclease activity.
- Gaps are filled by DNA pol I by adding dNTPs.
- The gap between the two DNA fragments is sealed by DNA ligase, which helps in the formation of phosphodiester bonds.
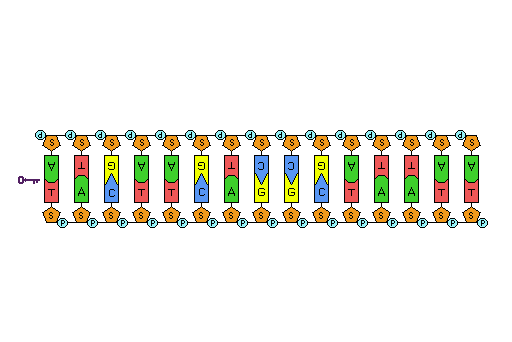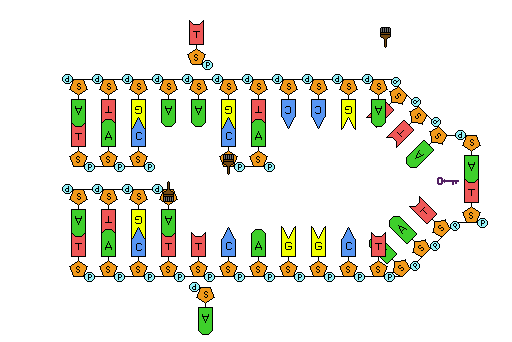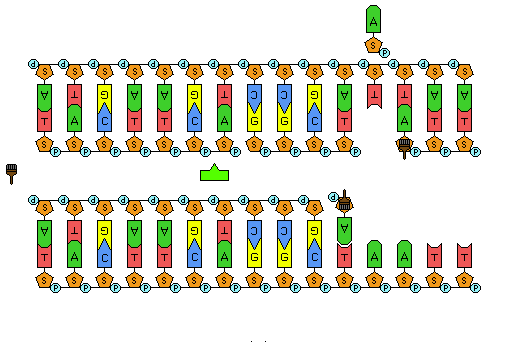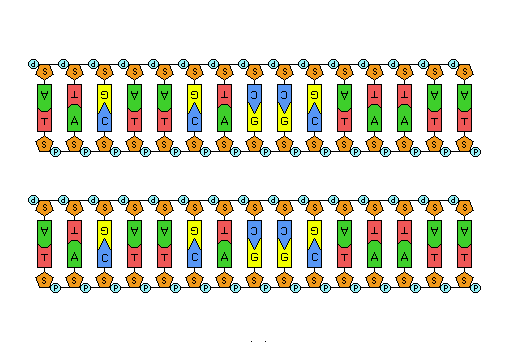
Figure 7: Animation of DNA replication. The double stranded DNA is separated. DNA pol III builds DNA complementary to the template strands (the strands that were separated). In this animation, the new strand on the bottom is the leading strand and is replicated continuously. In this animation, the new strand on the top is the lagging strand and is built in Okazaki fragments. DNA ligase repairs the nicks between the Okazaki fragments on the lagging strand to produce an unbroken DNA strand.
| Enzyme/protein | Specific Function |
|---|---|
| DNA pol I | Removes RNA primer and replaces it with newly synthesized DNA |
| DNA pol III | Main enzyme that adds nucleotides in the 5'-3' direction |
| Helicase | Opens the DNA helix by breaking hydrogen bonds between the nitrogenous bases |
| Ligase | Seals the gaps between the Okazaki fragments to create one continuous DNA strand |
| Primase | Synthesizes RNA primers needed to start replication |
| Sliding Clamp | Helps to hold the DNA polymerase in place when nucleotides are being added |
| Topoisomerase | Helps relieve the strain on DNA when unwinding by causing breaks, and then resealing the DNA |
| Single-strand binding proteins (SSB) | Binds to single-stranded DNA to prevent DNA from rewinding back. |
Laboratory Instructions

Figure 8: Puzzle pieces that can be used to build DNA and RNA.
Procedure:
DNA Structure & RNA Structure
- Organize the puzzle pieces so that all of the puzzle pieces and in piles by type (e.g. one pile for phosphate groups, one pile for deoxyribose, one pile for guanine, etc.).
- Answer questions 1-5 in the "Results & Questions" section under "DNA Structure."
- Build eight DNA bases: one with adenine, one with thymine, one with guanine, and one with cytosine.
- Build four RNA bases: one with adenine, one with uracil, one with guanine, and one with cytosine.
- Answer questions 6-8 in the "Results & Questions" section under "DNA Structure."
- Connect DNA nucleotides to each other to form the sequence: 5'-GTAC-3'
- Connect RNA nucleotides to each other to form the sequence: 5'-GUAC-3'
- Answer questions 9-11 in the "Results & Questions" section under "DNA Structure."
- Build a complementary strand of DNA to the DNA sequence already built and pair them together.
- Answer questions 12-17 in the "Results & Questions" section under "DNA Structure."
DNA Replication
- Build the following DNA nucleotides:
- 4 adenine DNA nucleotides
- 4 thymine DNA nucleotides
- 8 guanine DNA nucleotides
- 8 cytosine DNA nucleotides
- Build a single strand of DNA with the following structure: 5'-AGCCTG-3'
- Build the complementary DNA strand to the sequence above and pair it with the DNA molecule you made in step 2.
- Answer questions 1-6 in the "Results & Questions" section under "DNA Replication."
- The segment of DNA you built is the origin of replication. Presto-change-o! You are now the enzyme helicase! Do what helicase would do.
- Answer questions 7-9 in the "Results & Questions" section under "DNA Replication."
- Presto-change-o! You are now the enzyme DNA pol III (a DNA polymerase the elongates growing DNA strands during DNA replication). Build new DNA molecules to complement the template DNA strands starting at the 5' end and ending at the 3' end of the new strands (just like DNA pol III would do).
- Answer questions 10-13 in the "Results & Questions" section under "DNA Replication."
- Disassemble all of the puzzle pieces and return to the box/bag where they came from.
LAB ASSIGNMENT
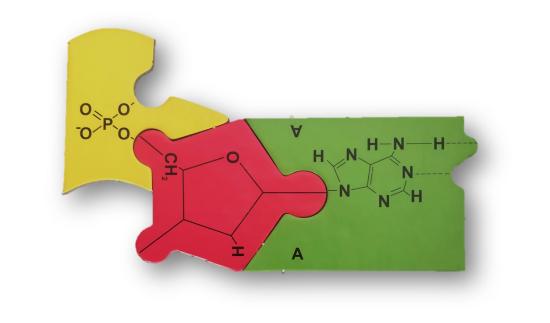
To be answered during scheduled lab time:
- DNA nucleotides contain deoxyribose as the pentose (5-carbon) sugar and RNA nucleotides contain ribose as the pentose sugar. Compare the chemical structures written on the deoxyribose and ribose puzzle pieces. What is the difference between ribose and deoxyribose?
- Consider your answer to question 1. What do you suppose "deoxy-" is referring to in the name "deoxyribose?"
- Identify the different base puzzle pieces (there are 5 types). Examine the chemical structures of the bases written on these puzzle pieces. Why do you suppose that DNA and RNA bases are often referred to as "nitrogenous bases?"
- Adenine and guanine are classified as "purines" and thymine, cytosine, and uracil are classified "pyrimidines." Looking at the chemical structures written on these puzzle pieces, what pattern do you notice that distinguishes purines from pyrimidines?
- Adenine pairs with thymine in DNA and with uracil in RNA. Guanine pairs with cytosine in both DNA and RNA. What generalization can you make about base-pairing and whether bases are purines or pyrimidines (e.g. do purines pair with purines, to pyrimidines pair with pyrimidines, or do purines pair with pyrimidines)?
- Compare the the DNA nucleotides with the RNA nucleotides. What similarities are there?
- Compare the the DNA nucleotides with the RNA nucleotides. What differences are there?
- On the image above showing a DNA nucleotide, write in 5' to show the 5' side of the nucleotide and write in 3' to show the 3' side of the nucleotide.
- Covalent bonds were formed between the nucleotides you joined together. These covalent bonds have a special name. What are these bonds called?
- Identify the 5' end and the 3' end of the DNA strand. Identify the 5' end of the RNA strand and the 3' end of the DNA strand. What component of the nucleotides do you find at the 5' end of these strands?
- What is missing from the DNA puzzle?
- Compare the DNA and RNA structures. What new difference is apparent in their structures?
- Examine the two DNA strands paired together. Notice that one DNA strand faces 5' to 3' and the other DNA strand is in the reverse direction. What is this arrangement called?
- The two DNA strands can easily be pulled apart from each other at the bases (unlike other locations in the puzzle, such as between the phosphates and the bases). Why is this? What types of interactions hold the bases of two DNA strands together?
- What are hydrogen bonds?
- How many hydrogen bonds are formed in the A-T pair?
- How many hydrogen bonds are formed in the G-C pair?
DNA Replication
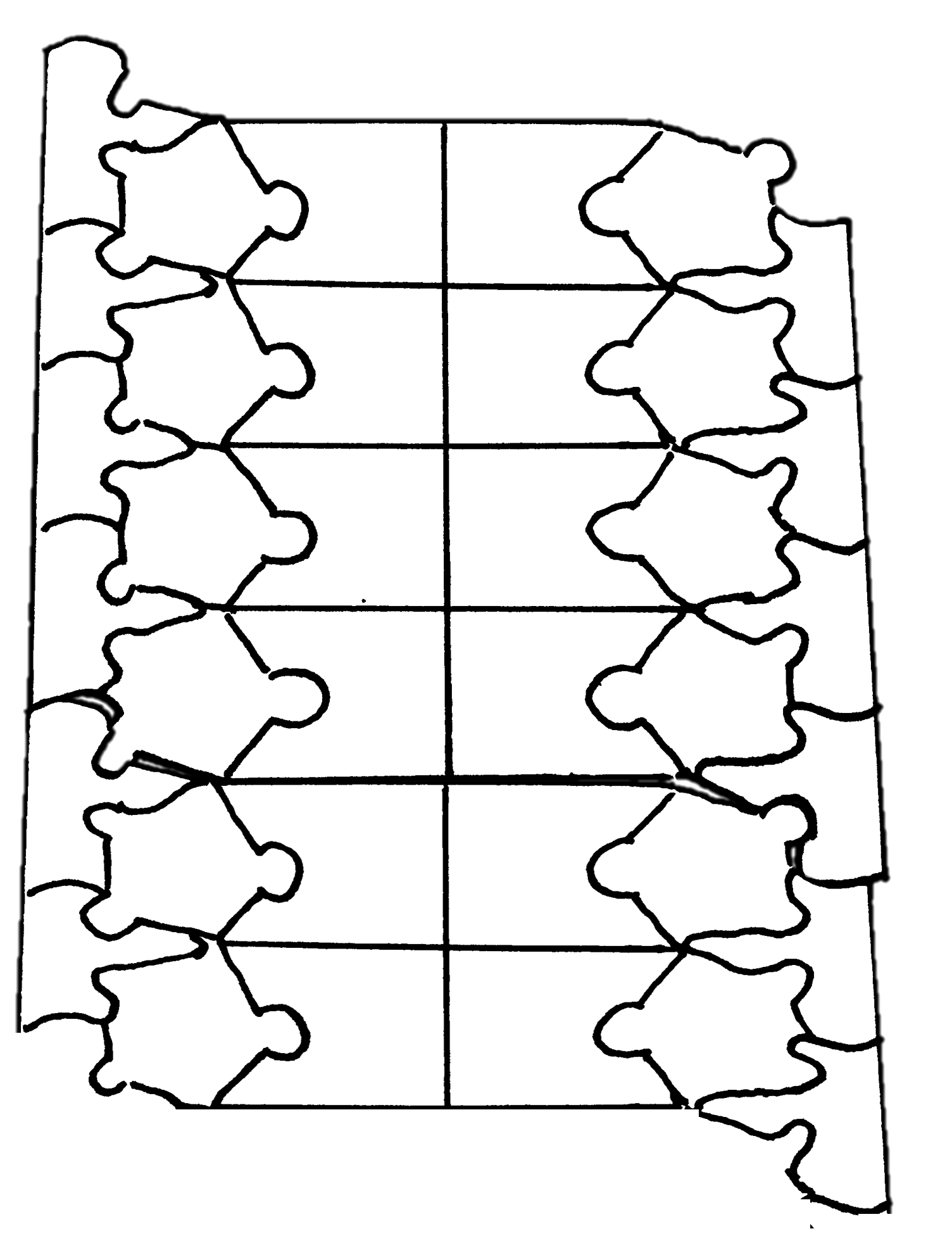
- Fill in the diagram above with the following:
- phos. = phosphate group
- deoxy. = deoxyribose
- A = adenine
- T = thymine
- G = guanine
- C = cytoskne
- 5' - show the 5' end on both DNA strands
- 3' - show the 3' end on both DNA strands
- The two DNA strands interact at the bases with hydrogen bonds. Are hydrogen bonds weak or strong attractions?
- How might it be possible for the two DNA strands to separate at the bases?
- What is the purpose of DNA replication?
- Where on a DNA molecule does DNA replication begin?
- What DNA replication enzyme breaks the hydrogen bonds between the nitrogenous bases to create single-stranded sections of DNA?
- What did helicase do to the double-stranded DNA puzzle?
- What enzyme elongates a growing DNA strand during DNA replication?
- What end of a growing DNA molecule can the enzyme named in question 8 add new nucleotides to (5' or 3')?
- What features of the actual process of DNA replication are missing from this re-creation of DNA replication? There are at least three.
- Would DNA pol III be able to create a new DNA molecule such as we did in this puzzle re-creation of DNA replication? Explain your answer.
- How is DNA replication a semiconservative process and how did we show this in the puzzle re-creation of DNA replication?
- Compare the newly replicated DNA double-strands in your puzzle pieces. Closely examine the sequences of the DNA and the directions of the strands. Are the two double strands identical? Give the DNA sequences with 5' and 3' directionality to explain.
Attributions
- Biology 2e by OpenStax is licensed under CC BY 4.0
- Circular bacterial chromosome replication.gif by Catherinea228 is licensed under CC BY-SA 4.0
- Cover Image: DNA animation.gif by brian0918™ is in the public domain
- Difference DNA RNA-EN.svg by File:Difference DNA RNA-DE.svg: Sponk / *translation: Sponk is licensed under CC BY-SA 3.0
- Kuensting replication color.gif by Steven Kuensting is licensed under CC BY-SA 4.0


