8: Plaque Assay and Biochemical Tests (Day 1)
- Page ID
- 105497
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Describe methods of viral quantification.
- Execute proper use of a micropipet.
- Successfully perform and interpret results of the plaque assay.
- Identify the importance of special media in culturing and identifying microorganisms.
- Describe the differences between selective, differential, and enrichment media.
- Compare various types of special media, including their mechanisms.
- Identify the appropriate special media to use.
- Interpret growth results on special media.
Quantification of Viruses
How do we count viruses?
Unlike bacteria, many of which can be grown on an artificial nutrient medium, viruses require a living host cell for replication. Infected host cells (eukaryotic or prokaryotic) can be cultured and grown, and then the growth medium can be harvested as a source of virus. In this lab, we will be quantifying bacteriophages (viruses that attack bacteria) using a plaque assay. This means that we must grow both bacteria and virus.
As the virus replicates and grows, it will lyse bacteria resulting in an area of clearing that can be observed on a bacterial lawn. These clearings are called plaques. A plaque assay, therefore, is a method by which the number of plaques observed can be used to estimate the amount of virus present in the original culture. The technique required to do so is called the pour-plate technique because you will mix the virus with the bacteria with melted agar and then pour the mixture onto a plate.
The unit of viral titer (concentration) is plaque-forming units per milliliter (pfu/ml).
As we do not know the starting concentration of virus, we will need to do a serial dilution (similar to the standard plate count). After incubation, the number of plaques formed will be used to calculate the original viral (phage) titer.
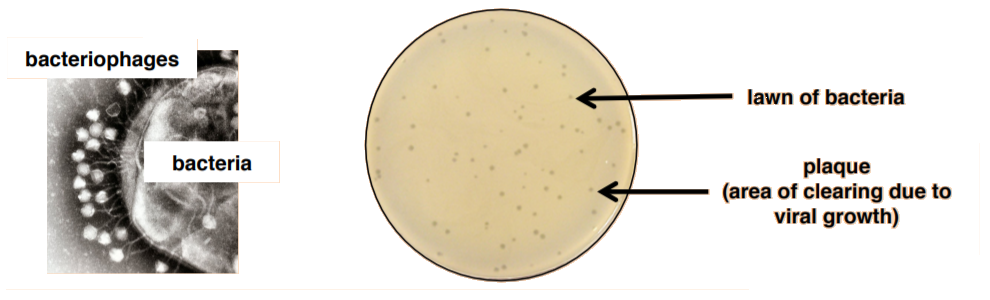
Plaque Assay:
Complete the Following Lab with Your Partner:
1. Obtain 14 microcentrifuge tubes and label 7 tubes 1-7 and 7 tubes E. coli 1-7.
2. Obtain 7 nutrient agar plates. Label them 1-7 and place them in the 37°C incubator to warm up until needed.
Dilution of Enterobacteria phage T4
Note
Enterobacteria phage T4 is a bacteriophage that infects Escherichia coli bacteria.
3. Aseptically add 990 μL of sterile saline to tube #1.
4. Aseptically add 900 μL of sterile saline to tubes #2-7.
5. Finger vortex the E. coli broth. Then, aseptically add 300 μL to all seven E. coli microcentrifuge tubes.
6. Vortex the T4 bacteriophage suspension and then aseptically transfer 10 μL to tube #1. Close the lid and vortex. This is your 10-2 dilution.
7. Aseptically transfer 100 μL from tube #1 to tube #2, close the lid and vortex. This is your 10-3 dilution.
8. Aseptically transfer 100 μL from tube #2 to tube #3, close the lid and vortex. This is your 10-4 dilution.
9. Aseptically transfer 100 μL from tube #3 to tube #4, close the lid and vortex. This is your 10-5 dilution.
10. Aseptically transfer 100 μL from tube #4 to tube #5, close the lid and vortex. This is your 10-6 dilution.
11. Aseptically transfer 100 μL from tube #5 to tube #6, close the lid and vortex. This is your 10-7 dilution.
12. Aseptically transfer 100 μL from tube #6 to tube #7, close the lid and vortex. This is your 10-8 dilution.
Addition of diluted phage to E. coli
13. Aseptically transfer 100 μL of SALINE into tube E. coli #1, close the lid and vortex. This will become your negative control.
14. Aseptically transfer 100 μL of dilution tube #2 to E. coli #2, close the lid and vortex.
15. Aseptically transfer 100 μL of dilution tube #3 to E. coli #3, close the lid and vortex.
16. Aseptically transfer 100 μL of dilution tube #4 to E. coli #4, close the lid and vortex.
17. Aseptically transfer 100 μL of dilution tube #5 to E. coli #5, close the lid and vortex.
18. Aseptically transfer 100 μL of dilution tube #6 to E. coli #6, close the lid and vortex.
19. Aseptically transfer 100 μL of dilution tube #7 to E. coli #7, close the lid and vortex.
20. Let all seven tubes stand for 15 minutes. We are allowing time for the virus to infect the bacteria.
21. Remove one soft agar tube (containing 2.5 ml of agar) from the hot water bath and add the entire contents of E. coli #1 to it. Gently mix.
22. Immediately pour the mixture onto plate A. Then, gently tilt the plate back and forth until the soft agar mixture is spread evenly across the solid medium.
Note
Remember that you need to take the soft agar plates and nutrient agar plates out one at a time. Otherwise, the agar will solidify before you have a chance to pour.
23. Allow the agar to solidify completely with the lid on at your table (15-20 minutes). Then, invert and place in the 37°C incubator to grow overnight.
24. Repeat 21-23 for the remaining dilutions of bacteriophage.
Biochemical Test (Day 1):
Today we will be learning about the use of special media for identification and enrichment of bacteria. We will begin with some basics, including the difference between selective, differential, and enrichment media. Additionally, we will learn the mechanism behind these special medias by exploring the composition of each special media. Lastly, we will learn how to decide which media best suits our microbiological needs. After completing this lab, you should be able to bring together the things you've learned about special media and how they are used to identify microorganisms discussed in Exam 3 material.
Microbes are EVERYWHERE...so how can we isolate only the specimen we want?
I'm sure your first thought was, "Duh, we dilute the specimen by streak- or spread-plate techniques." BUT, what if the specimen you are looking for is only a small fraction of the total microbes in your sample? If we perform dilution techniques, we may lose sight of our desired microbe completely. Special media techniques take advantage of microbial nutritional and environmental requirements to enrich your specimen of interest. Additionally, special medias can be used to select or differentiate various microbes, thus allowing for identification.
General Purpose Media
This is the media we most commonly use in lab. General purpose media will typically support the growth of a wide diversity of microorganisms. Examples of general purpose media include nutrient agar and tryptic soy agar (see table below). While this media is not special, it's important you understand how special media differs from general purpose media.
| Type of General Purpose Media | Components |
| Nutrient Agar |
|
| Tryptic Soy Agar |
|
As you can see, both types of general purpose media contain similar components. Agar for solidification, various digests or extracts for nutrients, and sodium chloride (salt) to maintain appropriate tonicity.
Enrichment Media
Enrichment media is used to culture fastidious microbes that grow slowly or are found in low abundance. The enrichment culture contains specific and/or essential nutrients which allow for increased growth of the microbe of interest. For example, some microbes may require blood or lysed red blood cells, while others require unique growth factors or vitamins. It is important to note that enrichment media DOES NOT inhibit growth of other microbes; it merely promotes growth of a specific microorganism. As a result, enrichment cultures do not always result in a pure culture. Instead, you may observe multiple colony types with your microbe of interest being more abundant. Common examples of enrichment media include sheep blood agar and chocolate agar.
Selective Media
Selective media contains ingredients that prevent the growth of unwanted microbes without inhibiting the growth of the desired microorganism. Most commonly, selective media takes advantage of the microbes unique characteristics. For example, say your microbe of interest can grow at a low pH (acid). Most microbes prefer a neutral pH and will not grow on acidic media. Consequently, by using acidic media we select for our microbe of interest by inhibiting the growth of unwanted microbes. An example of selective media is Mannitol Salt Agar (which is also differential). We will explore this media further in the next module and in today's lab assignment. Note that physical conditions can also be used for selection, including temperature.
Mannitol Salt Agar
| Ingredients |
|
Differential Media
Differential media contains ingredients that allow you to distinguish between organisms growing on the sample plate. The basis for differentiation is usually a biochemical process that reacts with specific indicators in the media, resulting in a color change. It's important to note that differential media alone does not select for any one organism, but instead allows for identification of different microbes. One example we just learned about was blood agar. In lecture we discussed different types of hemolysis or lysis of red blood cells. The resulting lysis (or absence of lysis) provides a visual indicator, allowing for differentiation of different microbes. Let's revisit blood agar:
Right away you should notice that there are three different microbes growing on this plate. We know this because we see three different types of hemolysis: beta hemolysis, alpha hemolysis, and gamma hemolysis. Recall that beta hemolysis is complete lysis (yellow appearance), alpha hemolysis is partial lysis (yellow-brown appearance), and gamma hemolysis is the absence of lysis (no color change).
Summary
Cultivation of Media for Bacteria, by Cindy Arvidson
To further explore different types of special media, we will be exploring various types of special media using resources created by Cindy Arvidson. This resource includes everything we just went over, as well as numerous examples of special media and how different microbes grow on special media. Please familiarize yourself with the composition and use of the special media listed in the provided resource. Additionally, familiarize yourself with how different microbes grow on special media by clicking the "Examples" link under each type of special media. You can even click on the individual example pictures to further observe colony morphology.
Procedure
- Obtain a liquid culture of a bacterial culture and Gram stain the specimen, then inoculate the following forms of media:
- NOTE: The above links should serve as a guide for inoculation of media. You will only be inoculating one microbe onto each type of media. The oxidase test will be performed on Day 2 of Biochemical Testing, as will any additional steps required for visualization of differential media.
- Today you will inoculate Starch Agar, SIM Agar, Mannitol Salt Agar, and EMB Agar.
LAB ASSIGNMENT
Record your results below. A countable plate has between 30-300 plaques. If there are more than 300 plaques record TMTC (too many to count), if there are fewer than 30 plaques record TFTC (too few to count) (1).
| A | B | C | D | E | F | G | |
| Plaques Counted | |||||||
| Sample Volume | 2.5 mL. | 2.5 mL. | 2.5 mL. | 2.5 mL. | 2.5 mL. | 2.5 mL. | 2.5 mL. |
| \(\frac{PFU}{mL}\) | |||||||
| \(\frac{PFU}{mL\;of\;starting\;tube}\) |
1. Why is it important in this assay to make sure that there are enough bacteria present to produce a lawn?
2. What would happen if you had not waited the 15 minutes that the protocol requires?
3. Why was the water bath set to 50°C?
4. What is the difference between a CFU and a PFU?

