9: Biochemical Tests (Day 2)
- Page ID
- 105499
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)BIOCHEMICAL TESTS (DAY 2):
CATALASE TEST:
1. On a glass slide, add 1-2 drops of H2O2.
2. Add loopful of a single species of bacteria from your plate to the slide; bubbles = positive result
OXIDASE TEST: 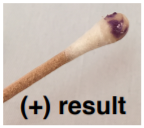
1. Swipe bacteria from your assigned PLATE onto a cotton swab.
2. Dispense 1-2 drops of the oxidase reagent onto the swab and record result.
STARCH PLATES:
1. Add enough iodine reagent to flood your plate. WAIT 5 MINUTES. The presence of clear halos surrounding colonies is positive for their ability to digest the starch and indicates the presence of alpha-amylase.
SIM MEDIA:
1. Add 5 drops of Kovac's reagent to the tube to detect indole production.
Biochemical Test Index
MANNITOL SALT AGAR PLATE (MSA):
Selective for gram-positive bacteria (e.g. Staphylococcus and Micrococcus). Mannitol fermentation by pathogenic staphylococci, such as S. aureus, is indicated by the media changing to yellow.
SELECTIVE AGENT: NaCl (salt)
DIFFERENTIAL AGENT: mannitol sugar fermentation
INDICATOR: Phenol red

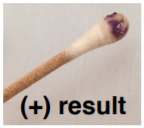 OXIDASE TEST:
OXIDASE TEST:
Used to determine if a bacterium has enzyme cytochrome oxidase. The final stage of bacterial respiration involves a series of membrane-embedded components collectively known as the electron transport chain. The final step in the chain may involve the use of the enzyme cytochrome oxidase, which catalyzes the oxidation of cytochrome c while reducing oxygen to form water. The reagent is usually N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) or N,N-dimethyl-pphenylenediamine (DMPD), which is also a redox indicator. The reagent is colorless if negative, the color turns blue/purple when oxidized.
SIM MEDIUM:
For the differentiation of gram-negative enteric bacilli on the basis of sulfide production, indole formation, and motility. H2S production is detected when ferrous sulfide, a black precipitate, is produced as a result of ferrous ammonium sulfate reacting with H2S gas. Casein peptone is rich in tryptophan, bacteria that possess the enzyme tryptophanase degrade tryptophan to indole. Indole is detected upon the addition of Kovacs Reagent producing a red band at the top of the medium. The semi-solid agar allows for the detection of bacterial motility. Motile organisms extend from the stab line and produce turbidity or cloudiness throughout the medium. Non-motile organisms grow only along the stab line and leave the surrounding medium clear.
| Sulfur | - | + | + | - |
| Indole | + | - | - | - |
| Motility | - | + | - | - |
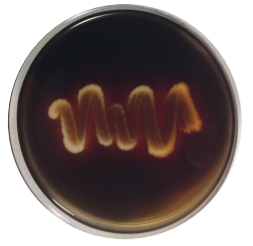 STARCH AGAR:
STARCH AGAR:
Starch is too large to pass through the plasma membrane and must be split into individual glucose molecules. Bacteria that have the exoenzyme amylase are able to hydrolyze starch by secreting these enzymes into the environment around them. After bacteria are allowed to grow, iodine is added to detect the presence of starch. Iodine complexes with starch to form a blue-black color in the culture medium. Clear halos surrounding colonies is indicative of their ability to digest the starch and results in a zone of clearance around plated growth. (+) = halo (-) = no halo
SELECTIVE AGENTS: none
DIFFERENTIAL AGENT: starch
INDICATOR: iodine
EMB AGAR:
LAB ASSIGNMENT
1. Report your results using the table below:
| Specimen: | |
| TSA slant | |
| Starch Agar | |
| SIM deeps | |
| MSA | |
| EMB Agar |
2. Interpret your results. In other words, what do your results say about the microorganisms metabolism?
| Specimen: | |
| TSA slant | |
| Starch Agar | |
| SIM deeps | |
| MSA | |
| EMB Agar |
3. How did the results observed on the MSA and EMB agar correlate to the Gram reaction of the bacteria?
4. What is the importance of biochemical tests in microbiology?
5. Why is it important to have a monoculture before you start biochemical testing?


