11.5: Enzyme Regulation
- Page ID
- 140313
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Regulation of Enzymes
Enzymes play a crucial role in cellular processes, and their regulation is essential for maintaining proper metabolic function. Cells have developed several sophisticated mechanisms to control enzyme activity, allowing them to respond quickly to changing conditions and metabolic needs.
Feedback Inhibition
Molecules can regulate enzyme function in many ways. The major question remains, however: What are these molecules and where do they come from? Some are cofactors and coenzymes, as you have learned. What other molecules in the cell provide enzymatic regulation such as allosteric modulation, and competitive and non-competitive inhibition? Perhaps the most relevant sources of regulatory molecules, with respect to enzymatic cellular metabolism, are the products of the cellular metabolic reactions themselves. In a most efficient and elegant way, cells have evolved to use the products of their own reactions for feedback inhibition of enzyme activity. Feedback inhibition involves the use of a reaction product to regulate its own further production (Figure \(\PageIndex{1}\)). The cell responds to an abundance of the products by slowing down production during anabolic or catabolic reactions. Such reaction products may inhibit the enzymes that catalyzed their production through the mechanisms described above.
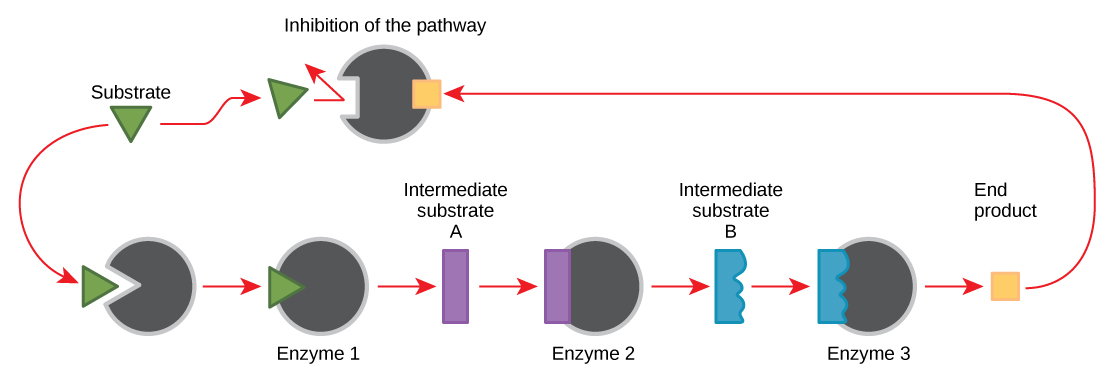
The production of both amino acids and nucleotides is controlled through feedback inhibition. Additionally, ATP is an allosteric regulator of some of the enzymes involved in the catabolic breakdown of sugar, the process that creates ATP. In this way, when ATP is in abundant supply, the cell can prevent the production of ATP. On the other hand, ADP serves as a positive allosteric regulator (an allosteric activator) for some of the same enzymes that are inhibited by ATP. Thus, when relative levels of ADP are high compared to ATP, the cell is triggered to produce more ATP through sugar catabolism.
Competitive and Non-Competitive Inhibitors
Competitive and noncompetitive inhibitors are two crucial mechanisms cells employ to regulate enzyme activity. Competitive inhibitors function by competing with the substrate for the enzyme's active site. These inhibitors structurally resemble the enzyme's substrate and bind reversibly to the active site, effectively preventing the substrate from binding. Importantly, competitive inhibition can be overcome by increasing substrate concentration, as this raises the probability of the substrate out-competing the inhibitor for the active site.
In contrast, noncompetitive inhibitors, also known as allosteric inhibitors, regulate enzymes by binding to a site other than the active site, called the allosteric site. This binding causes a conformational change in the enzyme, altering the shape of the active site and making it less effective. Another key difference is that noncompetitive inhibition cannot be overcome by increasing substrate concentration, as the inhibitor doesn't directly compete with the substrate for the active site.
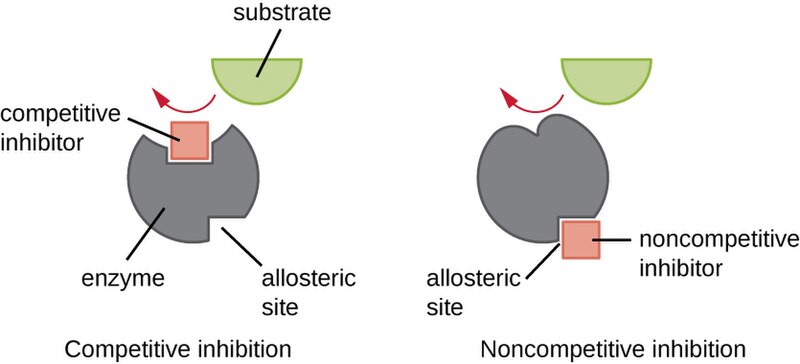
Both types of inhibition play significant roles in cellular regulation. Competitive inhibition often serves as a rapid, reversible way to control metabolic pathways, while noncompetitive inhibition allows for more complex regulation, as the allosteric site can respond to molecules unrelated to the substrate. Understanding these mechanisms is essential for comprehending how cells fine-tune their metabolic processes and respond to changing conditions. This knowledge also has important implications in drug development, as many pharmaceuticals work by inhibiting specific enzymes. By utilizing these inhibition mechanisms, cells can precisely control enzymatic activity to maintain homeostasis and adapt to varying metabolic demands.
The short video below provides an animation of how competitive and noncompetitive inhibitors regulate enzymes.
Allosteric Regulation
One of the primary ways cells regulate enzymes is through allosteric regulation. This process involves regulatory molecules, called allosteric effectors, binding to specific sites on the enzyme that are separate from the active site. When these effectors bind, they cause shape changes in the enzyme that can either activate or inhibit its function. This allows for rapid and reversible control of enzyme activity based on the cell's current needs.
Protein Modifications
Another important regulatory mechanism is covalent modification. In this process, chemical groups such as phosphates are added to or removed from specific amino acids in the enzyme. Phosphorylation, the addition of a phosphate group, is a particularly common form of covalent modification. These modifications can activate or inhibit the enzyme, providing a way for cells to quickly adjust enzyme activity in response to various signals.
Environmental Factors
Environmental factors also play a role in enzyme regulation. Changes in pH, temperature, or substrate concentration can affect an enzyme's shape and efficiency. Cells can use these environmental influences to their advantage, fine-tuning enzyme activity based on current conditions.
Localization
In eukaryotic cells, compartmentalization serves as another regulatory mechanism. By segregating enzymes into specific organelles or cellular compartments, cells ensure that enzymes only encounter their substrates in appropriate locations, providing an additional layer of control over metabolic processes.
Gene Regulation
Lastly, cells can regulate enzyme levels through genetic mechanisms. By controlling gene expression, cells can induce or repress the synthesis of specific enzymes based on their current needs. This allows for long-term adjustments in the quantity of enzymes available within the cell.

