11.4: Enzymes
- Page ID
- 140312
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
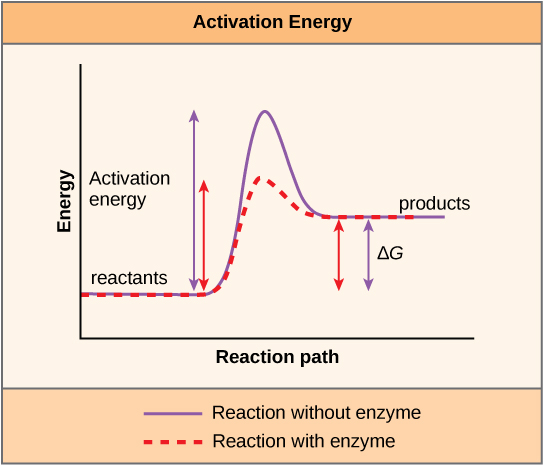
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)A substance that helps a chemical reaction to occur is called a catalyst, and the molecules that catalyze biochemical reactions are called enzymes. Most enzymes are proteins and perform the critical task of lowering the activation energies of chemical reactions inside the cell. Most of the reactions critical to a living cell happen too slowly at normal temperatures to be of any use to the cell. Without enzymes to speed up these reactions, life could not persist. Enzymes do this by binding to the reactant molecules and holding them in such a way as to make the chemical bond-breaking and -forming processes take place more easily. It is important to remember that enzymes do not change whether a reaction is exergonic (spontaneous) or endergonic. This is because they do not change the free energy of the reactants or products. They only reduce the activation energy required for the reaction to go forward (Figure \(\PageIndex{1}\)). In addition, an enzyme itself is unchanged by the reaction it catalyzes. Once one reaction has been catalyzed, the enzyme is able to participate in other reactions.
The chemical reactants to which an enzyme binds are called the enzyme’s substrates. There may be one or more substrates, depending on the particular chemical reaction. In some reactions, a single reactant substrate is broken down into multiple products. In others, two substrates may come together to create one larger molecule. Two reactants might also enter a reaction and both become modified, but they leave the reaction as two products. The location within the enzyme where the substrate binds is called the enzyme’s active site. The active site is where the “action” happens. Since enzymes are proteins, there is a unique combination of amino acid side chains within the active site. Each side chain is characterized by different properties. They can be large or small, weakly acidic or basic, hydrophilic or hydrophobic, positively or negatively charged, or neutral. The unique combination of side chains creates a very specific chemical environment within the active site. This specific environment is suited to bind to one specific chemical substrate (or substrates).
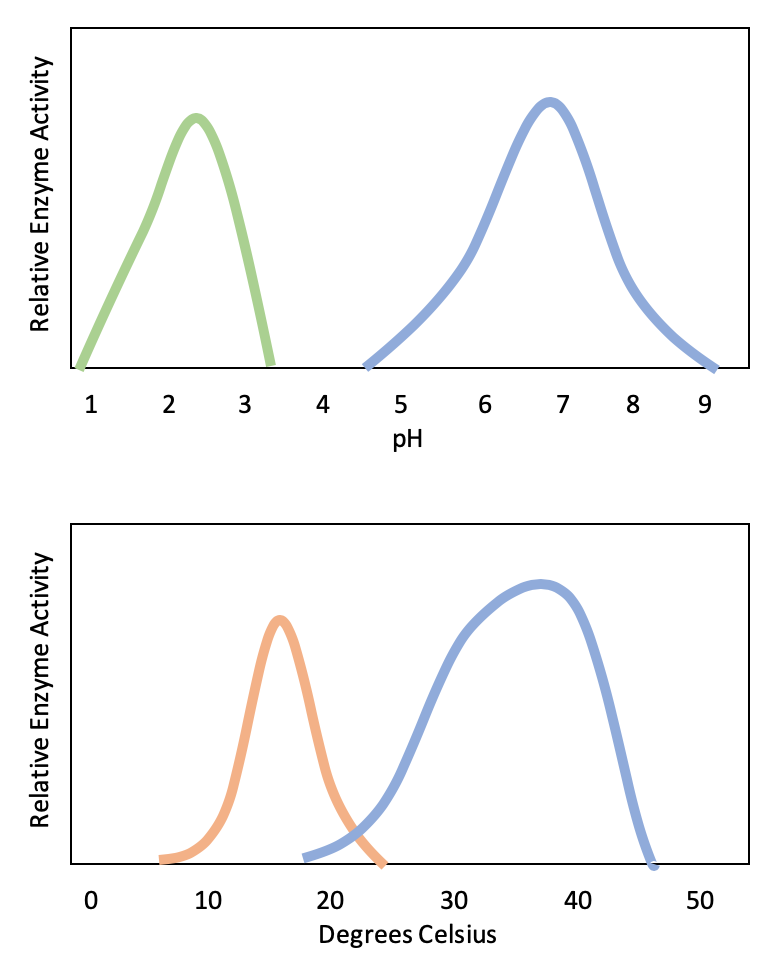
Active sites are subject to influences of the local environment. Increasing the environmental temperature generally increases reaction rates, enzyme-catalyzed or otherwise. However, temperatures outside of an optimal range reduce the rate at which an enzyme catalyzes a reaction. Hot temperatures will eventually cause enzymes to denature, an irreversible change in the three-dimensional shape and therefore the function of the enzyme (Figure \(\PageIndex{2}\)). Enzymes are also suited to function best within a certain pH and salt concentration range, and, as with temperature, extreme pH and salt concentrations can cause enzymes to denature.

Typically, enzymes function optimally in the environment where they are typically found and used. For example, the enzyme amylase is found in saliva, where it functions to break down starch (a polysaccharide – carbohydrate chain) into smaller sugars. Note that in this example, amylase is the enzyme, starch is the substrate, and smaller sugars are the product. The pH of saliva is typically between 6.2 and 7.6, with roughly 6.7 being the average. The optimum pH of amylase is between 6.7 and 7.0, which is close to neutral (Figure \(\PageIndex{3}\)). The optimum temperature for amylase is close to 37ºC (which is human body temperature).
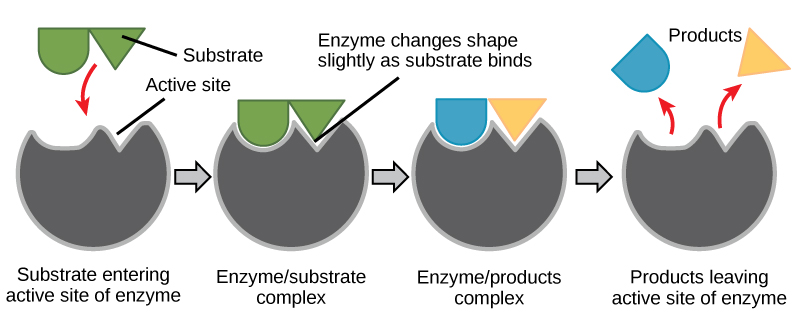
For many years, scientists thought that enzyme-substrate binding took place in a simple “lock and key” fashion. This model asserted that the enzyme and substrate fit together perfectly in one instantaneous step. However, current research supports a model called induced fit (Figure \(\PageIndex{4}\)). The induced-fit model expands on the lock-and-key model by describing a more dynamic binding between enzyme and substrate. As the enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that forms an ideal binding arrangement between enzyme and substrate.
When an enzyme binds its substrate, an enzyme-substrate complex is formed. This complex lowers the activation energy of the reaction and promotes its rapid progression in one of multiple possible ways.
- On a basic level, enzymes promote chemical reactions that involve more than one substrate by bringing the substrates together in an optimal orientation for reaction.
- Enzymes promote the reaction of their substrates by creating an optimal environment within the active site for the reaction to occur. The chemical properties that emerge from the particular arrangement of amino acid R groups (side chains) within an active site create the perfect environment for an enzyme’s specific substrates to react.
- The enzyme-substrate complex can also lower activation energy by compromising the bond structure so that it is easier to break.
- Finally, enzymes can also lower activation energies by taking part in the chemical reaction itself. In these cases, it is important to remember that the enzyme will always return to its original state by the completion of the reaction.
One of the hallmark properties of enzymes is that they remain ultimately unchanged by the reactions they catalyze. After an enzyme has catalyzed a reaction, it releases its product(s) and can catalyze a new reaction.
It would seem ideal to have a scenario in which all of an organism’s enzymes existed in abundant supply and functioned optimally under all cellular conditions, in all cells, at all times. However, a variety of mechanisms ensures that this does not happen. Cellular needs and conditions constantly vary from cell to cell, and change within individual cells over time. The required enzymes of stomach cells differ from those of fat storage cells, skin cells, blood cells, and nerve cells. Furthermore, a digestive organ cell works much harder to process and break down nutrients during the time that closely follows a meal compared with many hours after a meal. As these cellular demands and conditions vary, so must the amounts and functionality of different enzymes.
Query \(\PageIndex{1}\)
Query \(\PageIndex{2}\)
Query \(\PageIndex{3}\)
Query \(\PageIndex{4}\)
Query \(\PageIndex{5}\)
References
Unless otherwise noted, images on this page are licensed under CC-BY 4.0 by OpenStax.
Text adapted from: OpenStax, Concepts of Biology. OpenStax CNX. May 18, 2016 http://cnx.org/contents/b3c1e1d2-839...9a8aafbdd@9.10

