12.5.1: Procedures for Case Study #1
- Page ID
- 123444
MATERIALS
Gibson Oxidase Test Swab and either a plate of MacConkey agar or a plate of Cetrimide agar, and an EnterPluri-Test
PROCEDURE (to be done in groups of 3 )
Videos reviewing techniques used in this lab:
How to Chemically Fix a Microscope Slide with Methanol
How to Prepare a Slide for Staining when using Bacteria from an Agar Culture
A Review of the Critical Decolorization Step of the Gram Stain
Preliminary Tips for Using a Microscope
Focusing Using Oil Immersion (1000X) Microscopy
Keep in mind that organisms other than the Enterobacteriaceae and Pseudomonas can cause these infections, so in a real clinical situation other lab tests and cultures for bacteria other than those upon which this lab is based would also be done.
Flow chart of the tests you will be using today. See Fig. \(\PageIndex{1}\)

1. Perform a Gram stain on your unknown. Remember that the concentration of bacteria on slides prepared from taking bacteria off a petri plate tend to be much greater than those prepared by taking bacteria out of a broth culture, so be careful not to under decolorize. Continue decolorizing until the purple just stops flowing off of the bacterial smear, then wash with water.
2. If you have a Gram-negative bacillus, determine if it is a glucose fermenting Gram-negative bacillus like most Enterobacteriaceae or a glucose non-fermenting Gram-negative bacillus such as Pseudomonas by performing an oxidase test as follows:
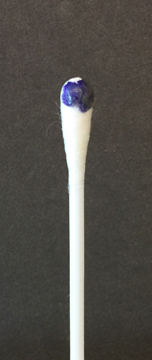
To perform the oxidase test, touch the oxidase test swab to a colony on your unknown plate. An oxidase-positive reaction will appear purple to black within 10 seconds as shown in Fig. \(\PageIndex{2A}\). An oxidase-negative reaction will result in no color change as shown in Fig. \(\PageIndex{2B}\). Ignore any color development occurring after 60 seconds.
Record your oxidase test results in the Oxidase test section of Lab 13.
|
Fig \(\PageIndex{2A}\): A Positive Oxidase Test |
Fig. \(\PageIndex{2B}\): A Negative Oxidase Test |
|---|---|
 |
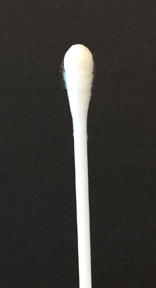 |
| Touch the Gibson Oxidase Test swab to a colony. An oxidase-positive reactions will appear purple to black within 10 seconds as shown in the above image. Disregard any color development that appears after 60 seconds. | Touch the Gibson Oxidase Test swab to a colony. An oxidase-positive reactions will appear purple to black within 10 seconds as shown in the above image. No change in color is negative. Disregard any color development that appears after 60 seconds. |
| Copyright; Gary E. Kaiser, Ph.D. The Community College of Baltimore County, Catonsville Campus CC-BY-3.0 | |
3. If your unknown is oxidase-negative, usually indicating a glucose fermenting Gram-negative bacillus, do the following inoculations:
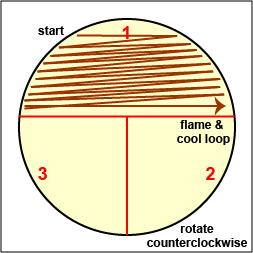
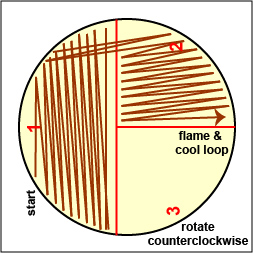
a. Streak your unknown for isolation on a plate of MacConkey agar, a selective medium used for the isolation of non-fastidious Gram-negative rods and particularly members of the family Enterobacteriaceae, as illustrated in Fig. \(\PageIndex{3A}\) through \(\PageIndex{3C}\). Incubate upside down and stacked in the petri plate holder on the shelf of the 37°C incubator corresponding to your lab section.
|
Fig \(\PageIndex{3A}\):Streaking For Isolation, Step-1 |
Fig. \(\PageIndex{3B}\): Streaking For Isolation, Step-2 |
Fig. \(\PageIndex{3C}\): Streaking For Isolation, Step-3 |
|---|---|---|
 |
 |
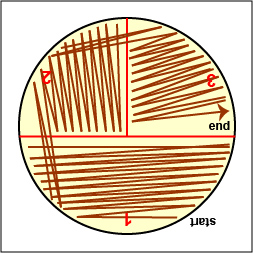 |
| Streak the inoculum over area "1" as shown above. Then flame the loop and cool it by sticking it into the edge of the agar. Rotate the plate 90 degrees counterclockwise. | Rotate the plate so area "1" is on your left. Drag your stetile inoculating loop through area "1" two times and spread out over area "2" as shown above. Then flame the loop and cool it by sticking it into the edge of the agar. Rotate the plate 90 degrees counterclockwise. | Rotate the plate so that area "2" is on your left. Drag your sterile inoculating loop through area "2" two times and spread the inoculum on area "2" over area "3." Sterilize the loop. |
| Copyright; Gary E. Kaiser, Ph.D. The Community College of Baltimore County, Catonsville Campus CC-BY-3.0 | ||

b. Inoculate an EnterPluri-Test as follows:
1. Remove both caps of the EnteroPluri-Test and with the straight end of the inoculating wire, pick off the equivalent of a colony from your unknown plate. A visible inoculum should be seen on the tip and side of the wire.
2. Inoculate the EnteroPluri-Test by grasping the bent-end of the inoculating wire, twisting it, and withdrawing the wire through all 12 compartments using a turning motion.
3. Reinsert the wire into the tube (use a turning motion) through all 12 compartments until the notch on the wire is aligned with the opening of the tube. (The tip of the wire should be seen in the citrate compartment.) Break the wire at the notch by bending. Do not discard the wire yet.
4. Using the broken off part of the wire, punch holes through the cellophane which covers the air inlets located on the rounded side of the last 8 compartments. Your instructor will show you their correct location. Discard the broken off wire in the disinfectant container.
5. Replace both caps and incubate the EnteroPluri-Test on its flat surface at 36°- 37°C for 18-24 hours.
4. If your unknown is oxidase-positive, usually indicating a glucose non-fermenting Gram-negative bacillus, do the following inoculation:
a. Streak your unknown for isolation on a plate of Cetrimide agar, a selective and differential medium for Pseudomonas, as illustrated in Fig. 9A through 9C above. Incubate upside down and stacked in the petri plate holder on the shelf of the 37°C incubator corresponding to your lab section.
Note that MacConkey agar can also be used to isolate Pseudomonas but we are using the Cetrimide agar today because it enables us to detect the production of the blue to green water-soluble pigment by Pseudomonas aeruginosa, as well as the production of fluorescein.
You will also inoculate an EnteroPluri-Test for practice only, but keep in mind that the EnteroPluri-Test is used to identify Enterobacteriaceae, not Pseudomonas.
Contributors and Attributions
Dr. Gary Kaiser (COMMUNITY COLLEGE OF BALTIMORE COUNTY, CATONSVILLE CAMPUS)

