12.3: Results
- Page ID
- 16029
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)A. ELISA test
Record the appearance of each of the wells here.
| Well | Contents | Color appearance | Interpretation |
| 1 | |||
| 2 | |||
| 3 | |||
| 4 | |||
| 5 | |||
| 6 |
B. Biolog
Record your Biolog results below.
Describe the colony that you picked for Biolog (color, etc.): _________________________________________ ________________________________________________________________
Use the sheet on the next page to record your results. In each box, write “+” if the well appears positive (purple color), “-” if there is no color change, and “+/-“ if the result is borderline (light purple).
Your instructor will show you how to enter your results into the database to achieve an identification.
An example of what a Biolog plate looks like is shown below.
Identification: ___________________________________
% match: _______________________________________
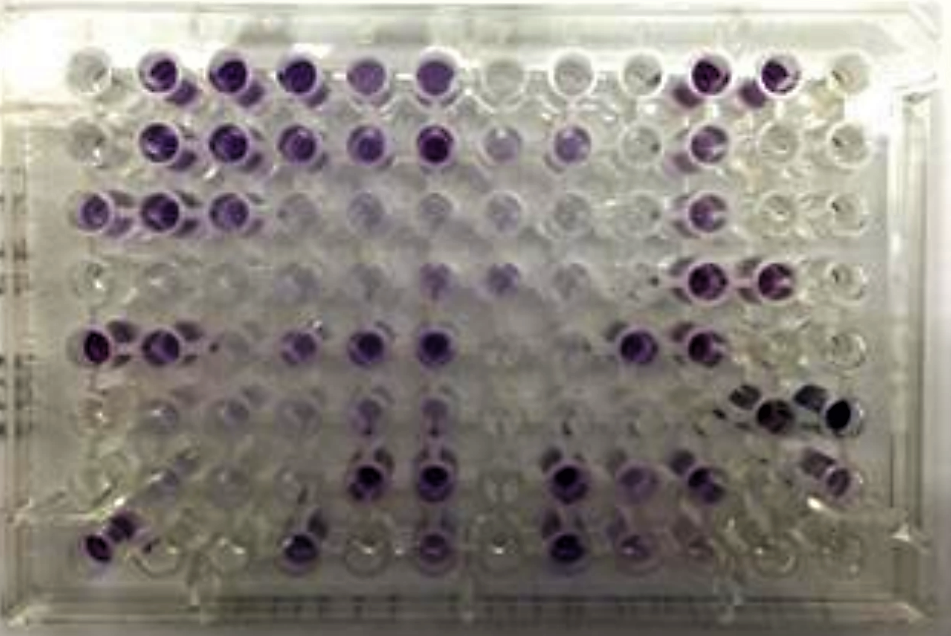.png?revision=1&size=bestfit&width=375&height=251)
| A1 Negative Control | A2 Dextrin | A3 D-Maltose | A4 D-Trehalose | A5 D-Cellobiose | A6 Gentiobiose | A7 Sucrose | A8 D-Turanose | A9 Stachyose | A10 Positive Control | A11 pH 6 | A12 pH 5 |
| B1 D-Raffinose | B2 α-D-Lactose | B3 D-Melibiose | B4 β-Methyl-D-Glucoside | B5 D-Salicin | B6 N-Acetyl-DGlucosamine | B7 N-Acetyl-β-D-Mannosamine | B8 N-Acetyl-D-Galactosamine | B9 N-Acetyl Neuraminic Acid | B10 1% NaCl | B11 4% NaCl | B12 8% NaCl |
| C1 α-D-Glucose | C2 D-Mannose | C3 D-Fructose | C4 D-Galactose | C5 3-Methyl Glucose | C6 D-Fucose | C7 L-Fucose | C8 L-Rhamnose | C9 Inosine | C10 1% Sodium Lactate | C11 Fusidic Acid | C12 D-Serine |
| D1 D-Sorbitol | D2 D-Mannitol | D3 D-Arabitol | D4 myo-Inositol | D5 Glycerol | D6 D-Glucose- 6- PO4 | D7 D-Fructose- 6- PO4 | D8 D-Aspartic Acid | D9 D-Serine | D10 Troleandomycin | D11 Rifamycin SV | D12 Minocycline |
| E1 Gelatin | E2 Glycyl-L-Proline | E3 L-Alanine | E4 L-Arginine | E5 L-Aspartic Acid | E6 L-Glutamic Acid | E7 L-Histidine | E8 L-Pyroglutamic Acid | E9 L-Serine | E10 Lincomycin | E11 Guanidine HCl | E12 Niaproof 4 |
| F1 Pectin Acid | F2 D-Galacturonic Acid | F3 L-Galactonic Acid Lactone | F4 D-Gluconic Acid | F5 D-Glucuronic Acid | F6 Glucuronamide | F7 Mucic Acid | F8 Quinic Acid | F9 D-Saccharic Acid | F10 Vancomycin | F11 Tetrazolium Violet | F12 Tetrazolium Blue |
| G1 p-Hydroxy Phenylacetic Acid | G2 Methyl Pyruvate | G3 D-Lactic Acid Methyl Ester | G4 L-Lactic Acid | G5 Citric Acid | G6 α-KetoGlutaric Acid | G7 D-Malic Acid | G8 L-Malic Acid | G9 BromoSuccinic Acid | G10 Nalidixic Acid | G11 Lithium Chloride | G12 Potassium Tellurite |
| H1 Tween 40 | H2 γ-aminobutyric Acid | H3 α- HydroxyButyric Acid | H4 β-HydroxyD,LButyric Acid | H5 α-Keto-Butyric Acid | H6 Acetoacetic Acid | H7 Propionic Acid | H8 Acetic Acid | H9 Formic Acid | H10 Aztreonam | H11 Sodium Butyrate | H12 Sodium Bromate |


