Lab 11: Biochemical Tests (Day 2)
( \newcommand{\kernel}{\mathrm{null}\,}\)
BIOCHEMICAL TESTS (DAY 2):
catalase test
Tests for the presence of catalase enzyme that converts hydrogen peroxide (H2O2) into water and oxygen gas
oxidase test
Used to determine if a bacterium has enzyme cytochrome oxidase. The reagent is colorless if negative and turns blue/purple when oxidized.
CATALASE TEST:
1. On a glass slide, add 1-2 drops of H2O2.
2. Add loopful of a single species of bacteria from your plate to the slide; bubbles = positive result
3. Repeat for all four bacteria AND YOUR ASSIGNED BACTERIA from your TSA plate
OXIDASE TEST: 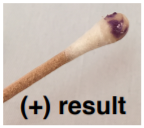
4. Swipe bacteria from your assigned PLATE onto a cotton swab.
5. Dispense 1-2 drops of the oxidase reagent onto the swab and record result.
6. Repeat for all four bacteria AND YOUR ASSIGNED BACTERIA from your TSA plate.
| Bacteria | Catalase +/- | Oxidase +/- |
| + | + | |
| + | - | |
| + | - | |
| - | - |
NITRATE BROTH:
Complete the following for each tube of the four tubes of nitrate broth. Observe the inverted Durham tube. If bubbles are present, then the following reaction has occurred and your bacteria is positive for nitrite and nitrate reductase:
NO3 (nitrate) → NO2 (nitrite) → N2 (gas)
1. If gas is NOT present, then add 3 drops of sufanilic acid (reagent A) and 3 drops of naphthlamine (reagent B) to your tube. Mix gently by tapping the bottom of the tube and wait 2-3 minutes. If your tube turns red, then bacteria is positive for nitrate reductase and the following reaction has occurred:
NO3 → NO2
2. If no red color is observed, add a TINY amount of powdered zinc using a toothpick and mix gently by tapping the bottom of the tube.
Note
If your sample turns red after step 2, DO NOT do step 3
3. Wait 15 minutes. If your tube turns red, then NO3 is still present and bacteria are nitrate reductase negative.
Note
Hint for lab practical – if I give you a red tube, how can you tell if it is nitrate positive or negative?
| NO3 present in the tube? | NO2 present in the tube? | N2 gas present in the tube? | |
| Assigned bacteria | |||
| Alcaligenes faecalis | |||
| Escherichia coli | |||
| Pseudomonas aeruginosa | |||
| NO3 is detected by? | NO2 is detected by? | N2 is detected by? | |
STARCH PLATES:
4. Add enough iodine reagent to flood your plate. WAIT 5 MINUTES. The presence of clear halos surrounding colonies is positive for their ability to digest the starch and indicates the presence of alpha-amylase.
GELATINASE TEST:
5. Place your gelatin tube in the ice bucket provided by the instructor for 10 minutes. Then, hold your tube sideways to determine if it is solid (negative result) or liquid (positive result). RECORD RESULTS FOR:
- CASEIN plate
- TRIBUTYRIN plate
- Phenol Red Broths
TSI SLANTS:
6. Record your results for your assigned bacteria using the table below.
| RESULTS (slant/butt) | SYMBOL | INTERPRETATION |
| Red/yellow | K/A | Glucose fermentation only; Peptone catabolized. |
| Yellow/yellow | A/A | Glucose and lactose and/or sucrose fermentation |
| Red/red | K/K | No fermentation; Peptone catabolized. |
| Red/no color change | K/NC | No fermentation; Peptone used aerobically |
| Yellow/yellow with bubbles | A/A,G | Glucose and lactose and/or sucrose fermentation; Gas produced. |
| Red/yellow with bubbles | K/A,G | Glucose fermentation only; Gas produced. |
| Red/yellow with bubbles and black precipitate | K/A,G,H2S | Glucose fermentation only; Gas produced; H2S produced. |
| Red/yellow with black precipitate | K/A,H2S | Glucose fermentation only; H2S produced. |
| Yellow/yellow with black precipitate | A/A, H2S | Glucose and lactose and/or sucrose fermentation; H2S produced. |
| No change/no change | NC/NC | No fermentation |
| A=acid production; G=gas production, K=alkaline reaction; H2S=sulfur reduction | ||
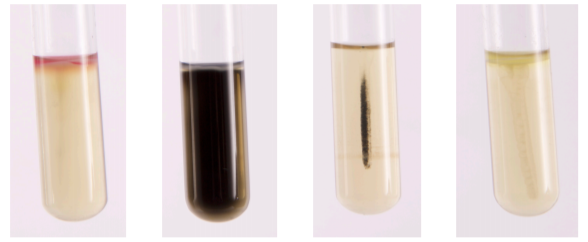
SIM MEDIA:
7. Add 5 drops of Kovacs reagent to the tube to detect indole production.
| Sulfur | - | + | + | - |
| Indole | + | - | - | - |
| Motility | - | + | - | - |
MR-VP:
8. Using a glass Pasteur pipet, take 1 ml out of your MR-VP broth tube and place it into a new tube
9. Add 5 drops MR methyl red reagent to the new tube, record results (positive result = red)
10. Using a glass pipet take 1 ml out of MR-VP broth tube and place it into a new tube
11. Add 6 drops VP-A and 3 drops VP-B, record results (positive result = red)
RECORD RESULTS FOR:
- Decarboxylate tubes
- Urea broth
- MAC plate
INDEX:
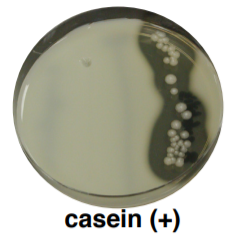 CASEIN (MILK) PLATE:
CASEIN (MILK) PLATE:
Casein is a large globular protein that gives milk its white and opaque color. It is too large to be transported across the cell membrane. Bacteria that have the exoenzyme 'casease' are able to hydrolyze casein by secreting this enzyme into the environment around them. Clear halos surrounding colonies are indicative of their ability to digest the casein and results in a zone of clearance around plated growth. (+) = halo (-) = no halo
SELECTIVE AGENT: none
DIFFERENTIAL AGENT: casein
INDICATOR: none
 CATALASE TEST:
CATALASE TEST:
Catalases are enzymes that convert hydrogen peroxide (H2O2) into water and oxygen gas. When a drop of peroxide is placed on catalase-producing bacteria, bubbles appear when the oxygen gas is formed. This is a test for aerobic (able to use oxygen) catalase-positive bacteria such as Staphylococcus and Micrococcus.
 CITRATE TEST:
CITRATE TEST:
Used for the detection of gram-negative enteric bacilli based on the ability of an organism to use citrate as the sole source of carbon and energy. Organisms capable of utilizing ammonium dihydrogen phosphate and citrate will grow unrestricted on this medium. Bromothymol blue acts as a pH indicator, causing the medium to change from green (neutral) to blue (alkaline) with increasing pH. Citrate utilization produces an alkaline carbonate, resulting in a deep blue color change within the agar. The medium will remain green if organisms do not grow or are not able to metabolize sodium citrate.
SELECTIVE AGENT: none
DIFFERENTIAL AGENT: ammonium dihydrogen phosphate and sodium citrate
INDICATOR: bromothymol blue
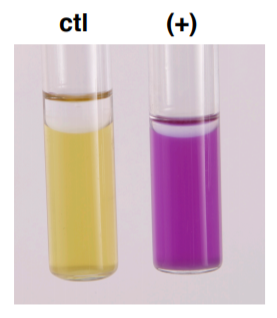 DECARBOXYLATE BROTHS:
DECARBOXYLATE BROTHS:
For the differentiation of gram-negative enteric bacilli based on their ability to break down specific amino acids. The production of the enzymes that break down amino acids (decarboxylase & dihydrolase enzymes) is induced in an acidic environment.
CONTROL: glucose fermented → acid → yellow
ARGININE, LYSINE or ORNITHINE: amino acid degraded → basic → purple
Microorganisms possessing the specific decarboxylase and dihydrolase enzymes for the amino acid (arginine, lysine or ornithine) degrade the amino acid to yield various amine by-products which create an alkaline environment that turns the indicator purplish. If the organism does not produce the appropriate enzyme, then the medium will remain yellow.
DIFFERENTIAL AGENT: Arginine, lysine or ornithine amino acids
INDICATOR: bromocresol purple and cresol red
 GELATIN HYDROLYSIS TEST:
GELATIN HYDROLYSIS TEST:
Nutrient gelatin is a differential medium that tests the ability of an organism to produce an exoenzyme, called gelatinase, that hydrolyzes gelatin. When gelatin is broken down, it can no longer solidify. If an organism can break down gelatin, the areas where the organism has grown will remain liquid even if the gelatin is chilled.
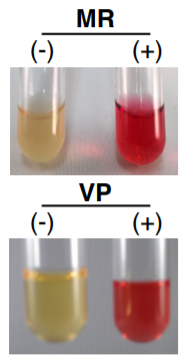 Methyl Red and Voges-Proskauer (MR-VP):
Methyl Red and Voges-Proskauer (MR-VP):
A medium that contains protein, glucose, and phosphate buffer. Some bacteria produce an acid while other bacteria further metabolize the acid to pH stable end products (Glucose→Acid→Stable End Product). The MR test is used to detect organisms capable of performing mixed acid fermentation where:
- Red=positive
- Orange=inconclusive
- Yellow=negative
The VP Test is designed for organisms who are able to ferment glucose to acids which are then converted to acetoin and 2,3 butanediol. The addition of VP reagents oxidizes acetoin to diacetyl and reacts with guanidine nuclei from the peptones to produce a red color. Remember:
- Red=positive results
- No color change or copper color=negative
MANNITOL SALT AGAR PLATE (MSA):
Selective for gram-positive bacteria (e.g. Staphylococcus and Micrococcus). Mannitol fermentation by pathogenic staphylococci, such as S. aureus, is indicated by the media changing to yellow.
SELECTIVE AGENT: NaCl (salt)
DIFFERENTIAL AGENT: mannitol sugar fermentation
INDICATOR: Phenol red

 NITRATE (NO3) BROTH:
NITRATE (NO3) BROTH:
Determines whether the microbe produces the enzymes nitrate reductase and nitrite reductase. The two enzymes catalyze two reactions involved in converting starting compound nitrate into end product nitrogen gas.
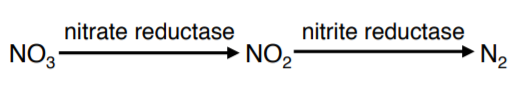
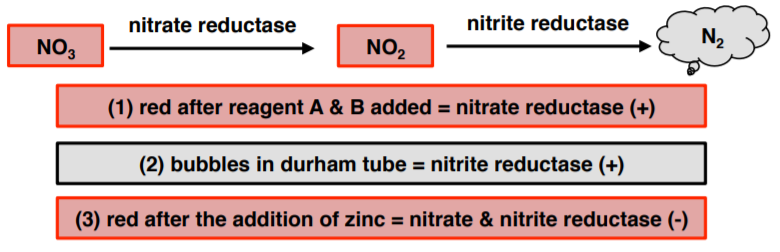
After incubating the nitrate broth, sulfanilic acid (reagent A or 1) and α-naphthylamine (reagent B or 2) are added. If a red-colored compound forms then nitrate reduction has occurred (NO3→NO2).
An inverted Durham tube tests for the presence of nitrogen gas (N2).
If neither a red color or gas is observed, then, confirmation is necessary that nitrate (NO3) remains in the broth. A SMALL addition of zinc dust will convert the nitrate to nitrite and form a red color. This test reaction is considered negative for nitrate reduction.
- Nitrate reductase (+) = red after reagent A & B added
- Nitrite and nitrate reductase (+) = bubbles in durham tube
- Nitrate & nitrite reductase (-) = red after the addition of zinc
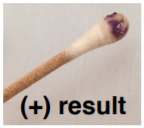 OXIDASE TEST:
OXIDASE TEST:
Used to determine if a bacterium has enzyme cytochrome oxidase. The final stage of bacterial respiration involves a series of membrane-embedded components collectively known as the electron transport chain. The final step in the chain may involve the use of the enzyme cytochrome oxidase, which catalyzes the oxidation of cytochrome c while reducing oxygen to form water. The reagent is usually N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) or N,N-dimethyl-pphenylenediamine (DMPD), which is also a redox indicator. The reagent is colorless if negative, the color turns blue/purple when oxidized.
 PHENOL RED BROTH:
PHENOL RED BROTH:
A general-purpose differential test medium typically used to differentiate gram-negative enteric bacteria. It contains peptone, phenol red (a pH indicator), one carbohydrate (i.e glucose or lactose), and a Durham tube to test for the ability to convert the end product of glycolysis, pyruvic acid, into CO2 gas.
DIFFERENTIAL AGENT: carbohydrate, Durham tube
INDICATOR: phenol red (-) (+)
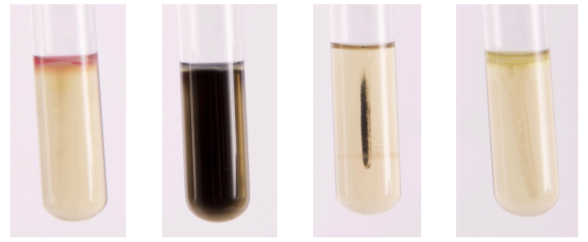
SIM MEDIUM:
For the differentiation of gram-negative enteric bacilli on the basis of sulfide production, indole formation, and motility. H2S production is detected when ferrous sulfide, a black precipitate, is produced as a result of ferrous ammonium sulfate reacting with H2S gas. Casein peptone is rich in tryptophan, bacteria that possess the enzyme tryptophanase degrade tryptophan to indole. Indole is detected upon the addition of Kovacs Reagent producing a red band at the top of the medium. The semi-solid agar allows for the detection of bacterial motility. Motile organisms extend from the stab line and produce turbidity or cloudiness throughout the medium. Non-motile organisms grow only along the stab line and leave the surrounding medium clear.
| Sulfur | - | + | + | - |
| Indole | + | - | - | - |
| Motility | - | + | - | - |
 STARCH AGAR:
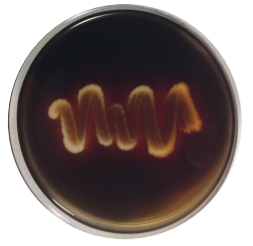
STARCH AGAR:
Starch is too large to pass through the plasma membrane and must be split into individual glucose molecules. Bacteria that have the exoenzyme amylase are able to hydrolyze starch by secreting these enzymes into the environment around them. After bacteria are allowed to grow, iodine is added to detect the presence of starch. Iodine complexes with starch to form a blue-black color in the culture medium. Clear halos surrounding colonies is indicative of their ability to digest the starch and results in a zone of clearance around plated growth. (+) = halo (-) = no halo
SELECTIVE AGENTS: none
DIFFERENTIAL AGENT: starch
INDICATOR: iodine
 TRIBUTYRIN AGAR:
TRIBUTYRIN AGAR:
Tributyrin oil is a type of lipid called a triglyceride. It is too large to be transported across the cell membrane. Bacteria that have the exoenzyme lipase are able to hydrolyze tributyrin oil and break it down. Tributyrin oil forms an opaque suspension in the agar. When an organism produces lipase and breaks down the tributyrin, a clear halo surrounds the areas where the lipase-producing organism has grown. (+) = halo (-) = no halo
SELECTIVE AGENTS: none
DIFFERENTIAL AGENT: tributyrin oil
INDICATOR: none
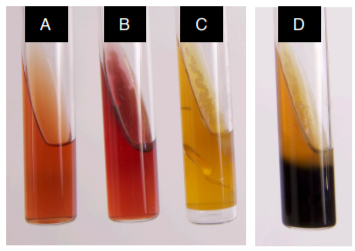
TRIPLE SUGAR IRON (TSI) SLANT/DEEP:
Tests the ability of bacteria to ferment sugars and to reduce sulfur to H2S, often used to identify Salmonella and Shigella. Medium contains sugars (lactose, sucrose, and glucose) and thiosulfate. Slant/deep allows for aerobic and anaerobic growth conditions. An alkaline/acid (red slant/yellow butt) reaction is indicative of glucose fermentation only. An acid/acid (yellow slant/yellow butt) reaction indicates the fermentation of glucose, lactose and/or sucrose. The absence of carbohydrate fermentation results in an alkaline/alkaline (red slant, red butt) reaction. If an organism can reduce sulfur, the hydrogen sulfide (H2S) gas which is produced will react with the ferrous ammonium sulfate ((NH4)2Fe(SO4)2 · 6H2O) to form iron sulfide (FeS), which appears as a black precipitate. If CO2 gas is produced, then the agar will be lifted or cracked as a result.
(A) Uninoculated
(B) K/K
(C) A/A+gas
(D) A/A+ H2S
Note
In (D), you can’t see the A because of the H2S
DIFFERENTIAL AGENT: glucose, lactose, sucrose, and sodium thiosulfate
INDICATOR: phenol red and ferrous ammonium sulfate
UREA BROTH:
Tests the ability of an organism to hydrolyze urea to ammonia and carbon dioxide using the enzyme urease. The broth contains urea and a very small amount of nutrients for the bacteria. The pH indicator is phenol red. If the urea in the broth is degraded and ammonia is produced, the pH rises and the media turns pink.
BACTERIAL UNKNOWN IDENTIFICATION:
You must work independently on this project! You will receive no help in staining, performing tests, interpreting tests, or even focusing the microscope other than your lab notebook and your flowchart. There should be NO conversations between students about their unknown experiment. You CAN NOT take pictures. I reserve the right to penalize students for giving or receiving unauthorized help from other students.
Each student will receive a broth culture of their unknown bacteria. The tube will be marked with a number only. Record this number and write it on all plates and tubes that you inoculate, along with your name and the date.
- Your unknown sample will contain 1 bacterial species.
- Do not discard any test results!
- You must request the media that you need from the instructor do not take anything without asking.
UNKNOWN LAB DAY 1:
- Gram stain your culture and record the results on DATA SHEET, and turn in your data sheet to the instructor (you will receive this document back at the beginning of day 2).
- Streak for isolated colonies on a TSA plate → this is a skills test.
- Streak for isolated colonies on a MAC.
- Store the original numbered unknown culture in your lab section’s rack at the front of the class.
UNKNOWN LAB DAY 2:
- Record the results of your MAC on DATA SHEET #1.
- Inoculate your chosen biochemical tests.
- You get 3 groups of tests for free from the list below. After that, each test will result in the loss of 5 points.
- EXO-ENZYMES (CASEIN AND TRYBUTERIN AND GEL)
- DECARBOXYLATE (LYSINE AND ORNITHINE)
- PHENOL RED BROTHS (GLUCOSE, LACTOSE, SUCROSE)
- OXIDASE
- TSI
- NITRATE
- UREA
- CITRATE
- MR-VP
- SIM
UNKNOWN LAB DAY 3:
- Record your test results on your DATA SHEET.
- Identify the GENUS and SPECIES of your unknown bacteria.
- Turn in DATA SHEET.



