9: Self assembly – Lipids, Membranes
- Page ID
- 6099
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Lecture 10. Self assembly – Lipids, Membranes
Reading & Problems: LNC p. 357-361, 362-364, 366-368, 368-370, 385-395 Figs. 11-17 and 11-18; p. 381 prob. 1, 2, 7, 9, 11, 12
I. Lipids - hydrophobic cellular "small" molecules - non-protein components of cells that partition into organic solvents like xylene or hexane.
A.  Fatty acids- long chain carboxylic acids; aphipathic (having both hydrophobic and hydrophilic regions on one molecule); salts of FA are soaps; form micelles in aqueous solutions(the structure of a micelle can be viewed at
Fatty acids- long chain carboxylic acids; aphipathic (having both hydrophobic and hydrophilic regions on one molecule); salts of FA are soaps; form micelles in aqueous solutions(the structure of a micelle can be viewed at  this site). Fatty acid properties are determined by chain length and number of unsaturations.
this site). Fatty acid properties are determined by chain length and number of unsaturations.
B.  Triglycerides (triacylglycerols) - three FA chains attached to a glycerol backbone via ester linkages. Called fats (if mostly saturated and solid at room temperature) or oils (if unsaturation leads them to be liquid at room temperature). Primarily used for energy storage.
Triglycerides (triacylglycerols) - three FA chains attached to a glycerol backbone via ester linkages. Called fats (if mostly saturated and solid at room temperature) or oils (if unsaturation leads them to be liquid at room temperature). Primarily used for energy storage.
C.  Glycerophospholipids - glycerol backbone with two FA chains and a phosphate (which may have additional functional groups attached). A major class of membrane lipids. Will spontaneously form bilayers or liposomes in aqueous environment.
Glycerophospholipids - glycerol backbone with two FA chains and a phosphate (which may have additional functional groups attached). A major class of membrane lipids. Will spontaneously form bilayers or liposomes in aqueous environment.
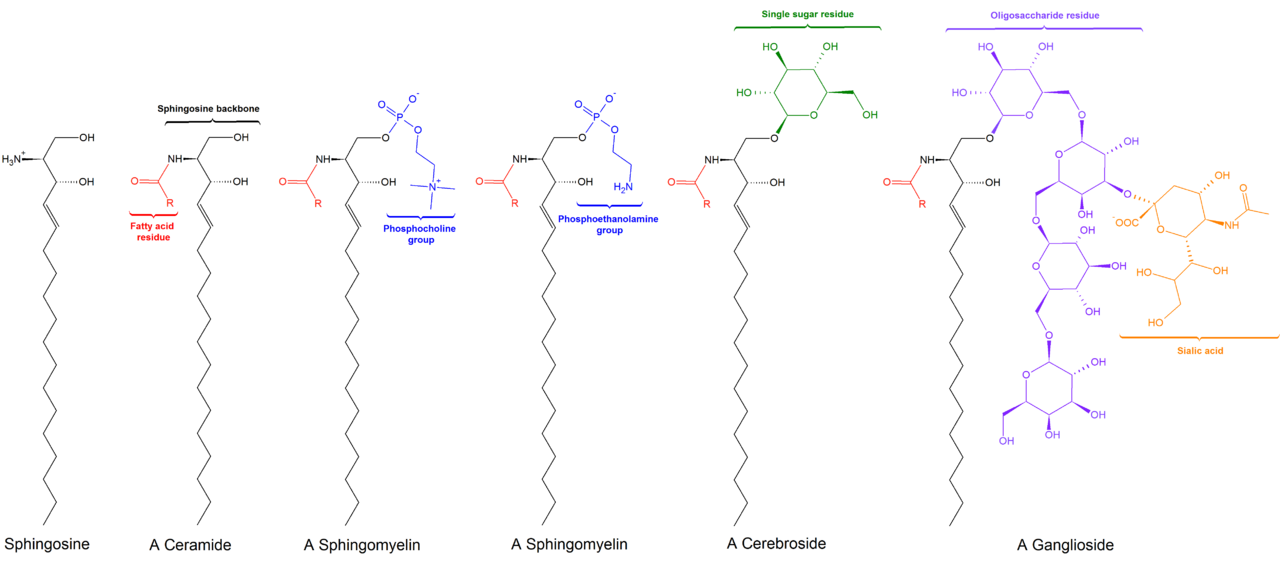
D. Sphingolipids - structures based on a sphingosine backbone. A major class of membrane lipids consist of a sphingospine backbone with a hydrophilic group attached to the backbone hydroxyl group and a FA attached to the backbone amino group via an amide linkage (peptide bond).  Sphingomyelin structure shows that it is similar to a
Sphingomyelin structure shows that it is similar to a  glycerophospholipid in overall shape. (Image Source: https://commons.wikimedia.org/wiki/F...structures.png)
glycerophospholipid in overall shape. (Image Source: https://commons.wikimedia.org/wiki/F...structures.png)
E.  Cholesterol. In membranes of animals. Broadens melting curve of membrane enabling maintenance of appropriate fluidity over a range of temperatures.
Cholesterol. In membranes of animals. Broadens melting curve of membrane enabling maintenance of appropriate fluidity over a range of temperatures.
II. Membranes - biological  lipid bilayers with associated/included proteins.(
lipid bilayers with associated/included proteins.( here's a simulation of motion in a lipid bilayer).
here's a simulation of motion in a lipid bilayer).
1. Integral membrane proteins - can only be removed from membrane by methods that lead to dissolution of the membrane. Surfaces of proteins in contact with interior of the membrane are hydrophobic.
2. Peripheral membrane proteins - can be removed by salt or other treatments that leave membrane largely intact. Can be bound to surface of integral membrane proteins, partially enter membrane, or be anchored by attached lipid molecules (e. g. prenylation).
Some take home information
Glycerolipids - have a glycerol backbone
Sphingolipids - have a sphingosine backbone
Phospholipids - include a phosphate group
Glycolipids - have attached sugar residues
Some Other Vocabulary: amphipathic, micelle, liposome, ester, vesicle, sonication, vortexing, ethanolamine, choline
Check out the structures that can be viewed under the "2. Lipids" heading on  this page, click on the "JSmol" button and you won't need Java to view the structures.
this page, click on the "JSmol" button and you won't need Java to view the structures.
Contributors
Charles S. Gasser (Department of Molecular & Cellular Biology; UC Davis)

