8: Protein Structure, Self assembly – Multimeric Proteins, Cytoskeletal fibers, Protein Evolution
( \newcommand{\kernel}{\mathrm{null}\,}\)
Reading:LNC p. 143-151, 104-108; Protein evolution problem set;  Protein evolution problems
Protein evolution problems
B. Final folded structure is minimum energy state.
Under physiological conditions folding of protein must have a negative delta G. The delta H for protein folding is strongly negative due to formation of hydrogen bonds and salt bridges in the folded state. The delta S for folding of the protein itself is very strongly negative due to the highly ordered state of the folded protein. However, the delta S for exclusion of water from the hydrophobic side chains that will be forced into the interior of the protein during folding is strongly positive. This contribution of a positive entropy change in the water largely (but not completely) offsets the negative entropy change for folding of the chain and enables the protein to fold. At higher temperatures, the entropy factor will predominate and the protein will unfold.
II. Multimeric proteins/quaternary structure - assembly of multiple subunits into a functional protein (Image Source: https://en.wikipedia.org/wiki/ATP_sy...p_synthase.PNG, https://commons.wikimedia.org/wiki/F...e-pdb-2ATC.png)
 Nucleosome
Nucleosome Mediator complex,
Mediator complex,  aspartate transcarbomylase trimer
aspartate transcarbomylase trimer
A. Protein subunits held together primarily by hydrophobic interactions, but also by salt bridges and H-bonds.
B. Contacting surfaces are complementary - hydrophobic patches will align with hydrophobic patches; H-bond donor will align with H-bond acceptor; positive charge will align with negative charge.
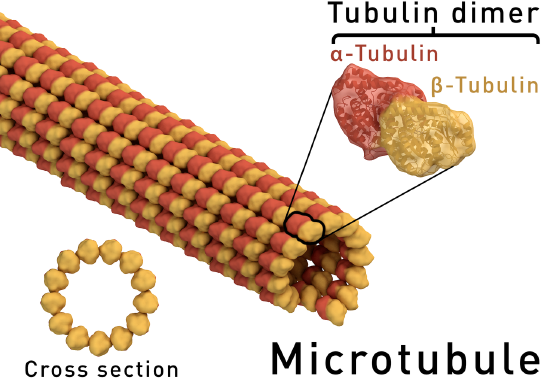
A. Example: Microtubules - polymers of alpha/beta tubulin dimers (Image Source: https://en.wikipedia.org/wiki/Microt..._structure.png)
B. Example: Actin filaments - polymers of actin. (image source: https://commons.wikimedia.org/wiki/F...ofilaments.jpg)
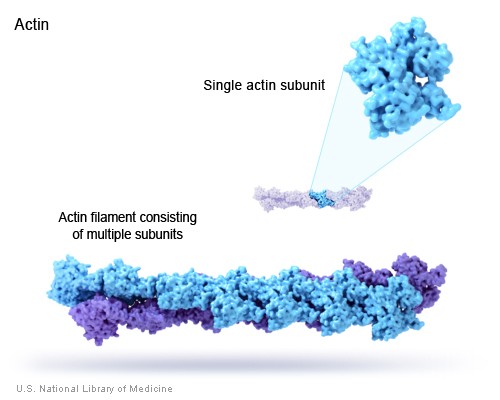
C. Principle of equivalence - in an assembly of identical subunits, all subunits must be in equivalent positions. Equivalence can be tested by seeing if a combination of rotation and translation can superpose any subunit on any other without changing the appearance of the assembly.
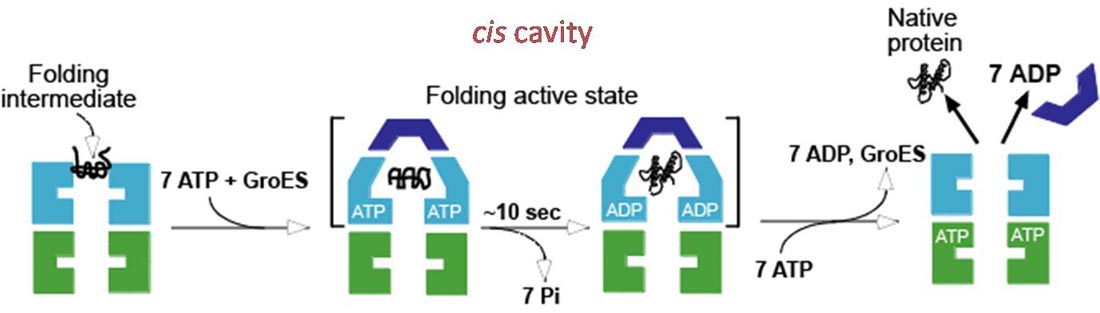
IV. Molecular Chaperones (Chaperonins) - protein complexes that help prevent improper folding or aggregation of nascent proteins.
A. Proteins in concentrated solutions (e. g. cytoplasm) can aggregate preventing proper folding, even though the folded state is the energy minimum.
B. Chaperonins can prevent aggregation and sequester unfolded proteins, or unassembled protein complexes and allow them time to fold/assemble into the energy minimizing state.
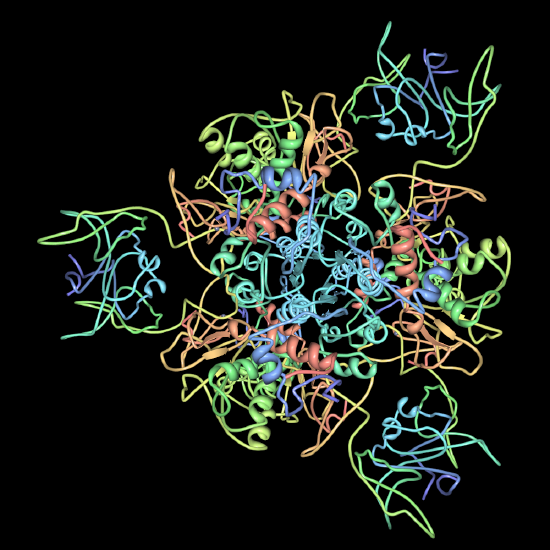
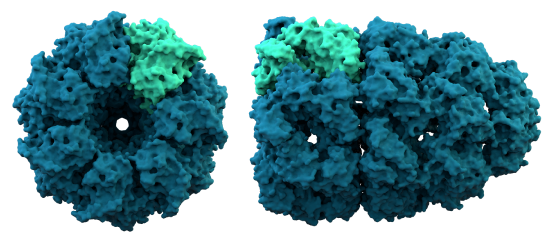
D. Chaperonins are large protein complexes and it appears that folding occurs in a central cavity. (image source: https://commons.wikimedia.org/wiki/F...ronin_1AON.png)
A. Relationships between proteins can be evaluated thorough protein alignments.  Globin alignment example.
Globin alignment example.
B. Orthologs are separated by a speciation event and are the "same gene" in two different species (e. g. mouse and human alpha globins) , paralogs are separated by a gene duplication event and represent two different genes in one species (e. g. mouse alpha globin and mouse beta globin). Homologs are any proteins related by descent from a common ancestor (orthologs and paralogs etc.)
Some take home information
ß-mercaptoethanol (BME) - a reducing agent that reduces (breaks) disulfide bonds, restoring the normal -SH groups on the cysteine side chains.
Urea, guanidine HCL - chaotropic agents that at high concentrations (e. g. 6 - 8 M) interfere with hydrophobic interactions (and to a lesser extent H-bonds) leading to unfolding of proteins.
Contributors
Charles S. Gasser (Department of Molecular & Cellular Biology; UC Davis)