Section 19.6: Chemical Defenses - Complement System
- Page ID
- 146388
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Plasma Protein Mediators
Many nonspecific innate immune factors are found in plasma, the fluid portion of blood. Plasma contains electrolytes, sugars, lipids, and proteins, each of which helps to maintain homeostasis (i.e., stable internal body functioning), and contains the proteins involved in the clotting of blood. Additional proteins found in blood plasma, such as acute-phase proteins, complement proteins, and cytokines, are involved in the nonspecific innate immune response.
There are two terms for the fluid portion of blood: plasma and serum. How do they differ if they are both fluid and lack cells? The fluid portion of blood left over after coagulation (blood cell clotting) has taken place is serum. Although molecules such as many vitamins, electrolytes, certain sugars, complement proteins, and antibodies are still present in serum, clotting factors are largely depleted. Plasma, conversely, still contains all the clotting elements. To obtain plasma from blood, an anticoagulant must be used to prevent clotting. Examples of anticoagulants include heparin and ethylene diamine tetraacetic acid (EDTA). Because clotting is inhibited, once obtained, the sample must be gently spun down in a centrifuge. The heavier, denser blood cells form a pellet at the bottom of a centrifuge tube, while the fluid plasma portion, which is lighter and less dense, remains above the cell pellet.
Acute-Phase Proteins
The acute-phase proteins are another class of antimicrobial mediators. Acute-phase proteins are primarily produced in the liver and secreted into the blood in response to inflammatory molecules from the immune system. Examples of acute-phase proteins include C-reactive protein, serum amyloid A, ferritin, transferrin, fibrinogen, and mannose-binding lectin. Each of these proteins has a different chemical structure and inhibits or destroys microbes in some way (Table \(\PageIndex{1}\)).
| Some Acute-Phase Proteins and Their Functions | |
|---|---|
| C-reactive protein | Coats bacteria (opsonization), preparing them for ingestion by phagocytes |
| Serum amyloid A | |
| Ferritin | Bind and sequester iron, thereby inhibiting the growth of pathogens |
| Transferrin | |
| Fibrinogen | Involved in formation of blood clots that trap bacterial pathogens |
| Mannose-binding lectin | Activates complement cascade |
The Complement System
The complement system is a group of plasma protein mediators that can act as an innate nonspecific defense while also serving to connect innate and adaptive immunity (discussed in the next chapter). The complement system is composed of more than 30 proteins (including C1 through C9) that normally circulate as precursor proteins in blood. These precursor proteins become activated when stimulated or triggered by a variety of factors, including the presence of microorganisms. Complement proteins are considered part of innate nonspecific immunity because they are always present in the blood and tissue fluids, allowing them to be activated quickly. Also, when activated through the alternative pathway (described later in this section), complement proteins target pathogens in a nonspecific manner.
The process by which circulating complement precursors become functional is called complement activation. This process is a cascade that can be triggered by one of three different mechanisms, known as the alternative, classical, and lectin pathways.
The alternative pathway is initiated by the spontaneous activation of the complement protein C3. The hydrolysis of C3 produces two products, C3a and C3b. When no invader microbes are present, C3b is very quickly degraded in a hydrolysis reaction using the water in the blood. However, if invading microbes are present, C3b attaches to the surface of these microbes. Once attached, C3b will recruit other complement proteins in a cascade (Figure \(\PageIndex{2}\)).
The classical pathway provides a more efficient mechanism of activating the complement cascade, but it depends upon the production of antibodies by the specific adaptive immune defenses. To initiate the classical pathway, a specific antibody must first bind to the pathogen to form an antibody-antigen complex. This activates the first protein in the complement cascade, the C1 complex. The C1 complex is a multipart protein complex, and each component participates in the full activation of the overall complex. Following recruitment and activation of the C1 complex, the remaining classical pathway complement proteins are recruited and activated in a cascading sequence (Figure \(\PageIndex{2}\)).
The lectin activation pathway is similar to the classical pathway, but it is triggered by the binding of mannose-binding lectin, an acute-phase protein, to carbohydrates on the microbial surface. Like other acute-phase proteins, lectins are produced by liver cells and are commonly upregulated in response to inflammatory signals received by the body during an infection (Figure \(\PageIndex{2}\)).
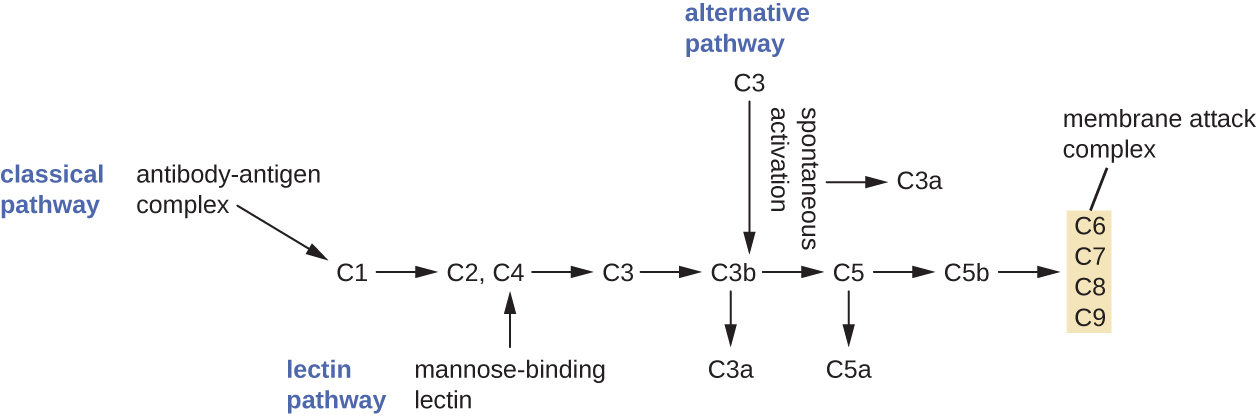
Although each complement activation pathway is initiated in a different way, they all provide the same protective outcomes: opsonization, inflammation, chemotaxis, and cytolysis. The term opsonization refers to the coating of a pathogen by a chemical substance (called an opsonin) that allows phagocytic cells to recognize, engulf, and destroy it more easily. Opsonins from the complement cascade include C1q, C3b, and C4b. Additional important opsonins include mannose-binding proteins and antibodies. The complement fragments C3a and C5a are well-characterized anaphylatoxins with potent proinflammatory functions. Anaphylatoxins activate mast cells, causing degranulation and the release of inflammatory chemical signals, including mediators that cause vasodilation and increased vascular permeability. C5a is also one of the most potent chemoattractants for neutrophils and other white blood cells, cellular defenses that will be discussed in the next section.
The complement proteins C6, C7, C8, and C9 assemble into a membrane attack complex (MAC), which allows C9 to polymerize into pores in the membranes of gram-negative bacteria. These pores allow water, ions, and other molecules to move freely in and out of the targeted cells, eventually leading to cell lysis and death of the pathogen (Figure \(\PageIndex{2}\)). However, the MAC is only effective against gram-negative bacteria; it cannot penetrate the thick layer of peptidoglycan associated with cell walls of gram-positive bacteria. Since the MAC does not pose a lethal threat to gram-positive bacterial pathogens, complement-mediated opsonization is more important for their clearance.
Key Concepts and Summary
- Numerous chemical mediators produced endogenously and exogenously exhibit nonspecific antimicrobial functions.
- Many chemical mediators are found in body fluids such as sebum, saliva, mucus, gastric and intestinal fluids, urine, tears, cerumen, and vaginal secretions.
- Antimicrobial peptides (AMPs) found on the skin and in other areas of the body are largely produced in response to the presence of pathogens. These include dermcidin, cathelicidin, defensins, histatins, and bacteriocins.
- Plasma contains various proteins that serve as chemical mediators, including acute-phase proteins, complement proteins, and cytokines.
- The complement system involves numerous precursor proteins that circulate in plasma. These proteins become activated in a cascading sequence in the presence of microbes, resulting in the opsonization of pathogens, chemoattraction of leukocytes, induction of inflammation, and cytolysis through the formation of a membrane attack complex (MAC).
- Cytokines are proteins that facilitate various nonspecific responses by innate immune cells, including production of other chemical mediators, cell proliferation, cell death, and differentiation.
- Cytokines play a key role in the inflammatory response, triggering production of inflammation-eliciting mediators such as acute-phase proteins, histamine, leukotrienes, prostaglandins, and bradykinin.


