2.4D: Ribosomes
- Page ID
- 3130
- Describe the structure and chemical composition of bacterial ribosomes and state their function.
- In terms of protein synthesis, briefly describe the process of transcription and translation.
- State, in a general sense, how antibiotics like neomycin, tetracycline, doxycycline, erythromycin, and azithromycin affect bacterial growth.
Ribosome Structure and Composition
Ribosomes are composed of ribosomal RNA (rRNA) and protein. Prokaryotic cells have three types of rRNA: 16S rRNA, 23S rRNA, and 5S rRNA. Like transfer RNA (tRNA), rRNAs use intrastrand H-bonding between complementary nucleotide bases to form complex folded structures. Ribosomes are composed of two subunits with densities of 50S and 30S ("S" refers to a unit of density called the Svedberg unit). The 30S subunit contains 16S rRNA and 21 proteins; the 50S subunit contains 5S and 23S rRNA and 31 proteins.The two subunits combine during protein synthesis to form a complete 70S ribosome about 25nm in diameter. A typical bacterium may have as many as 15,000 ribosomes.
Ribosomes are composed of two subunits that come together to translate messenger RNA (mRNA) into polypeptides and proteins during translation and are typically described in terms of their density. Density is the mass of a molecule or particle divided by its volume and is measured in Svedberg (S) units, a unit of density corresponding to the relative rate of sedimentation during ultra-high-speed centrifugation. The greater the S-value, the more dense the particle.
Ribosomal subunits are composed of ribosomal RNA (rRNA) and proteins. Ribosomal subunits with different S-values are composed of different molecules of rRNA, as well as different proteins. Remember that RNA is a polymer of ribonucleotides containing the nitrogenous base adenine, uracil, guanine, or cytosine. Different molecules of rRNA are of different lengths and have a different order of these ribonucleotide bases. Because rRNA is single stranded, many of the rRNA nucleotide bases are involved in intrastrand hydrogen bonds and this is what gives the rRNA molecule its specific shape (see Figure \(\PageIndex{1}\)). The shape, in turn, helps determine its function - much like the the interactions between amino acids in a protein determine that protein's shape and function (see Figure \(\PageIndex{2}\)).
Illustration of a 16S rRNA in Escherichia coli
Animation of a 16S rRNA
Illustration of the enzyme catalase
Prokaryotic ribosomes, for example, are composed of two subunits with densities of 50S and 30S. The 30S subunit contains 16S rRNA 1540 nucleotides long and 21 proteins; the 50S subunit contains a 5S rRNA 120 nucleotides long, a 23S rRNA 2900 nucleotides long, and 31 proteins. The two subunits combine during protein synthesis to form a complete 70S ribosome.
Eukaryotic ribosomal subunits have densities of 60S and 40S because they contain different rRNA molecules and proteins than prokaryotic ribosomal subunits. In most eukaryotes, the 40S subunit contains an 18S rRNA 1900 nucleotides long and approximately 33 proteins; the 60S subunit contains a 5S rRNA 120 nucleotides long, a 5.8S rRNA 160 nucleotides long, a 28S rRNA 4700 nucleotides long, and approximately 49 proteins. The two subunits combine during protein synthesis to form a complete 80S ribosome about 25nm in diameter.
Because of this difference in specific rRNAs and proteins the resulting "shape," there are drugs that can bind either to a 30S or 50S ribosomal subunit of a prokaryotic ribosome and subsequently block its function but are unable to bind to the equivalent 40S or 60S subunit of a eukaryotic ribosome.
Ribosome Functions
Ribosomes function as a workbench for protein synthesis, that is, they receive and translate genetic instructions for the formation of specific proteins. During protein synthesis, mRNA attaches to the 30s subunit and amino acid-carrying transfer RNAs (tRNA) attach to the 50s subunit (Figure \(\PageIndex{1}\)). Protein synthesis is discussed in detail in Unit 6.
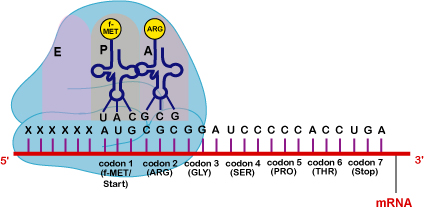
The chromosome is the genetic material of the bacterium. Genes located along the DNA are transcribed into RNA molecules, primarily messenger RNA (mRNA), transfer RNA (tRNA, and ribosomal RNA (rRNA). Messenger RNA is then translated into protein at the ribosomes.
- Transcription: Ribonucleic acid (RNA) is synthesized by complementary base pairing of ribonucleotides with deoxyribonucleotides to match a portion of one strand of DNA called a gene. Although genes are present on both strands of DNA, only one strand is transcribed for any given gene. Following transcription of genes into mRNA, 30S and 50S ribosomal subunits attach to the mRNA and tRNA inserts the correct amino acids which are subsequently joined to form a polypeptide or a protein through a process called translation.
- Translation: During translation, specific tRNA molecules pick up specific amino acids, transfer those amino acids to the ribosomes, and insert them in their proper place according to the mRNA "message." This is done by the anticodon portion of the tRNA molecules complementary base pairing with the codons along the mRNA.
In order for any of the tetracycline group of antibiotics to inhibit Gram-negative bacterial growth, they must enter the cytoplasm of that bacterium and bind to the 30S subunit of its ribosomes.
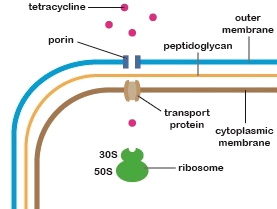
Earlier we learned the composition and functions of both the Gram-negative cell wall and the cytoplasmic membrane. We have also previously learned how the order of deoxyribonucleotide bases in DNA determines the order of ribonucleotide bases in rRNA which, in turn, determines the 3-dimensional shape of that RNA. Likewise, the order of deoxyribonucleotide bases in DNA determines the order of amino acids in a protein or enzyme which determines the 3-dimensional shape of that protein.
Considering all of this and using the illustration above, think of three physical changes that could occur within the bacterium as a result of acquiring new or altered genes through mutation or horizontal gene transfer that could enable the bacterium to resist that tetracycline.
Antimicrobial Agents that Alter Prokaryotic Ribosomal Subunits and Block Translation in Bacteria
Many antibiotics alter bacterial ribosomes, interfering with translation and thereby causing faulty protein synthesis. The portion of the ribosome to which the antibiotic binds determines how translation is effected. For example:
- The tetracyclines (tetracycline, doxycycline, demeclocycline, minocycline, etc.) bind reversibly to the 30S subunit, distorting it in such a way that the anticodons of charged tRNAs cannot align properly with the codons of the mRNA.
- The macrolides (erythromycin, azithromycin, clarithromycin, dirithromycin, troleandomycin, etc.) bind reversibly to the 50S subunit. They appear to inhibit elongation of the protein by preventing the enzyme peptidyltransferase from forming peptide bonds between the amino acids. They may also prevent the transfer of the peptidyl tRNA from the A-site to the P-site.
Antimicrobial chemotherapy will be discussed in greater detail later in Unit 2 under Control of Bacteria by Using Antibiotics and Disinfectants.
Summary
- Ribosomes are composed of ribosomal RNA (rRNA) and protein.
- Bacterial ribosomes are composed of two subunits with densities of 50S and 30S, as opposed to 60S and 40S in eukaryotic cells.
- Ribosomes function as a workbench for protein synthesis whereby they receive and translate genetic instructions for the formation of specific proteins.
- During translation, specific tRNA molecules pick up specific amino acids, transfer those amino acids to the ribosomes, and insert them in their proper place according to the mRNA "message."
- Many antibiotics bind to either the 30S or the 50S subunit of bacterial ribosomes, interfering with translation and thereby causing faulty protein synthesis.
Questions
Study the material in this section and then write out the answers to these questions. Do not just click on the answers and write them out. This will not test your understanding of this tutorial.
- Describe bacterial ribosomes. (ans)
- State the function of ribosomes. (ans)
- Define translation. (ans)
- State, in a general sense, how antibiotics like neomycin, tetracycline, doxycycline, erythromycin, and azithromycin affect bacterial growth. (ans)
- The tetracyclines (tetracycline, doxycycline) are antibiotics that bind to the 30S subunit of bacterial ribosomes. The macrolides (erythromycin, azithromycin, clarithromycin) are antibiotics that bind to the 50S subunit of bacterial ribosomes. Why won't these antibiotics be effective for fungal, protozoal, or viral infections? (ans)
- Multiple Choice (ans)


