14.1: Cell-Mediated Immunity - An Overview
- Page ID
- 3326
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Briefly compare humoral immunity with cell-mediated immunity.
- Define cell-mediated immunity and state what it is most effective against.
- State three different ways by which cell-mediated immunity protects the body.
- Define gene translocation and relate it to each T-lymphocyte being able to produce T-cell receptor with a unique shape.
- Define the following:
- combinatorial diversity
- junctional diversity
- In terms of cell-mediated immunity, state what is meant by anamnestic response and discuss its role in immune defense.
- Briefly describe why there is a heightened secondary response during anamestic response.
Cell-mediated immunity (CMI) is an immune response that does not involve antibodies but rather involves the activation of macrophages and NK-cells, the production of antigen-specific cytotoxic T-lymphocytes, and the release of various cytokines in response to an antigen . Cellular immunity protects the body by:
- Activating antigen-specific cytotoxic T-lymphocytes (CTLs) that are able to destroy body cells displaying epitopes of foreign antigen on their surface, such as virus-infected cells, cells with intracellular bacteria, and cancer cells displaying tumor antigens;
- Activating macrophages and NK cells, enabling them to destroy intracellular pathogens; and
- Stimulating cells to secrete a variety of cytokines that influence the function of other cells involved in adaptive immune responses and innate immune responses.
Cell-mediated immunity is directed primarily microbes that survive in phagocytes and microbes that infect non-phagocytic cells. It is most effective in destroying virus-infected cells, intracellular bacteria, and cancers. It also plays a major role in delayed transplant rejection.
Generation of T-cell receptor (TCR) diversity through gene translocation
As mentioned earlier, the immune system of the body has no idea as to what antigens it may eventually encounter. Therefore, it has evolved a system that possesses the capability of responding to any conceivable antigen. The immune system can do this because both B-lymphocytes and T-lymphocytes have evolved a unique system of gene-splicing called gene translocation, a type of gene-shuffling process where various different genes along a chromosome move and join with other genes from the chromosome. To demonstrate this gene translocation process, we will look at how each T-lymphocyte becomes genetically programmed to produce a T-cell receptor (TCR) having a unique shape to fit a specific epitope.
In a manner similar to B-lymphocytes, T-lymphocytes are able to cut out and splice together different combinations of genes along their chromosomes. Through random gene translocation, any combination of the multiple forms of each gene can join together. This is known as combinatorial diversity. The T-cell receptors or TCRs (Figure \(\PageIndex{1}\)) of most T-lymphocytes involved in adaptive immunity consist of an alpha (a) and a beta (ß) chain. There are 70-80 different Va genes and 61 different Ja genes that code for the variable portion of the a chain of the TCR. Likewise, there are 52 Vß genes, 1 Dß1 gene, 1 Dß2 gene, and 6-7 Jß genes that can recombine to form the variable portion of the TCR.
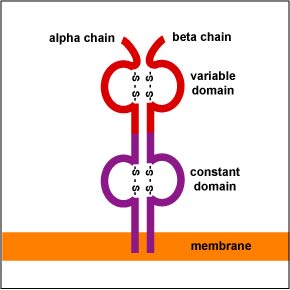
During gene translocation, specialized enzymes in the T-lymphocyte cause splicing inaccuracies wherein additional nucleotides are added or deleted at the various gene junctions. This change in the nucleotide base sequence generates even greater diversity in Fab shape. This is called junctional diversity. Unlike the BCR, somatic hypermutation does not occur during the production of the TCRs. As a result of combinatorial diversity and junctional diversity, each T-lymphocyte is able to produce a unique shaped T-cell receptor (TCR) capable of reacting with complementary-shaped peptide bound to a MHC molecule.
Anamnestic Response (Memory)
As a result of T-lymphocytes recognizing epitopes of protein antigens during cell-mediated immunity, numerous circulating T8-memory cells and T4-memory cells develop which possess anamnestic response or memory. These T-memory cells persist for the remainder of a person’s life. Effector memory T-cells (TEM cells) circulate in the blood whereas tissue resident memory T-cells (TRM cells) are found within the epithelium of the skin and mucous membranes. CD8 TRM cells are typically activated by viral antigens and subsequently produce inflammatory cytokines that trigger an innate immune response for nonspecific antiviral activity. CD4 TRM cells are found in clusters surrounding macrophages in the mucosa. Unlike TEM cells, TRM cells do not circulate in the blood and are not replenished from the blood. They remain in peripheral tissues.
A subsequent exposure to that same antigen results in:
- A more rapid and longer production of cytotoxic T-lymphocytes (CTLs);
- A more rapid and longer production of T4-effector lymphocytes; and
- Triggering of nonspecific innate immune responses.
Clonal Selection and Clonal Expansion
As mentioned above, during early differentiation of naive T-lymphocytes in the thymus marrow, each T4-lymphocyte and each T8-lymphocyte becomes genetically programmed to make a T-cell receptor or TCR with a unique shape through a series of gene translocations, and molecules of that TCR are put on its surface of that T-lymphocyte to function as its epitope receptor. When an antigen encounters the immune system, epitopes from protein antigens bound to MHC-I or MHC-II molecules eventually will react with a naive T4- and T8-lymphocyte with TCRs and CD4 or CD8 molecules on its surface that more or less fit and this activates that T-lymphocyte. This process is known as clonal selection.
Cytokines produced by effector T4-helper lymphocytes enable the now activated T4- and T8-lymphocyte to rapidly proliferate to produce large clones of thousands of identical T4- and T8-lymphocytes. In this way, even though only a few T-lymphocytes in the body may have TCR molecule able to fit a particular epitope, eventually many thousands of cells are produced with the right specificity. This is referred to as clonal expansion. These cells then differentiate into effector T4-lymphocytes and cytotoxic T-lymphocytes or CTLs.
Cellular immunity is also the mechanism behind delayed hypersensitivity (discussed later in this unit). Delayed hypersensitivity is generally used to refer to the harmful effects of cell-mediated immunity (tissue and transplant rejections, contact dermatitis, positive skin tests like the PPD test for tuberculosis, granuloma formation during tuberculosis and deep mycoses, and destruction of virus-infected cells).
Summary
- Cell-mediated immunity (CMI) is an immune response that does not involve antibodies but rather involves the activation of macrophages and NK-cells, the production of antigen-specific cytotoxic T-lymphocytes, and the release of various cytokines in response to an antigen.
- Cell-mediated immunity is directed primarily microbes that survive in phagocytes and microbes that infect non-phagocytic cells. It is most effective in destroying virus-infected cells, intracellular bacteria, and cancers.
- In a manner similar to B-lymphocytes, T-lymphocytes are able to randomly cut out and splice together different combinations of genes along their chromosomes through a process called gene translocation. This is known as combinatorial diversity and results in each T-lymphocyte generating a unique T-cell receptor (TCR).
- During gene translocation, specialized enzymes in the T-lymphocyte cause splicing inaccuracies wherein additional nucleotides are added or deleted at the various gene junctions. This change in the nucleotide base sequence generates even greater diversity in the shape of the TCR. This is called junctional diversity.
- As a result of combinatorial diversity and junctional diversity, each T-lymphocyte is able to produce a unique shaped T-cell receptor (TCR) capable of reacting with complementary-shaped peptide bound to a MHC molecule.
- As a result of T-lymphocytes recognizing epitopes of protein antigens during cell-mediated immunity, numerous circulating T8-memory cells and T4-memory cells) develop which possess anamnestic response or memory.
- A subsequent exposure to that same antigen results in a more rapid and longer production of cytotoxic T-lymphocytes (CTLs), and a more rapid and longer production of T4-effector lymphocytes.
- When an antigen encounters the immune system, epitopes from protein antigens bound to MHC-I or MHC-II molecules eventually will react with a naive T4- and T8-lymphocyte with TCRs and CD4 or CD8 molecules on its surface that more or less fit and this activates that T-lymphocyte. This process is known as clonal selection.
- Cytokines produced by effector T4-helper lymphocytes enable the now activated T4- and T8-lymphocyte to rapidly proliferate to produce large clones of thousands of identical T4- and T8-lymphocytes. This is referred to as clonal expansion.


