12.3: Oncogenes
- Page ID
- 5080
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)An oncogene is a gene that when mutated or expressed at abnormally-high levels contributes to converting a normal cell into a cancer cell. Cancer cells are cells that are engaged in uncontrolled mitosis.
The signals for normal mitosis
Normal cells growing in culture will not divide unless they are stimulated by one or more growth factors present in the culture medium (e.g, Epidermal Growth Factor (EGF)). The growth factor binds to its receptor, an integral membrane protein embedded in the plasma membrane with its ligand-binding site exposed at the surface of the cell. Examples:
- the Epidermal Growth Factor Receptor (EGFR). The gene encoding it, EGFR, is also known as HER1.
- another growth factor receptor is encoded by the gene ERBB2 (also known as HER2.)
- Binding of a growth factor to its receptor triggers a cascade of signaling events within the cytosol. Many of these involve
- kinases — enzymes that attach phosphate groups to other proteins. Examples: the proteins encoded by SRC, RAF, ABL, and the fusion protein encoded by BCR/ABL found in chronic myelogenous leukemia (CML).
- or molecules that turn on kinases. Example: RAS. RAS molecules reside on the inner surface of the plasma membrane where they serve to link receptor activation to "downstream" kinases like RAF.
- In most cases, phosphorylation activates the protein and eventually transfers the signal into the nucleus.
Here phosphorylation activates transcription factors that bind to promoters and enhancers in DNA, turning on their associated genes. Examples: AP-1, a heterodimer of the proteins encoded by jun and fos. Some of the genes turned on by these transcription factors encode other transcription factors (e.g., myc).
Some of the genes turned on by these downstream transcription factors encode cyclins that prepare the cell to undergo mitosis. Genes that participate in any one of the steps above can become oncogenes if they become mutated so that their product becomes constitutively active (that is, active all the time even in the absence of a positive signal) or they produce their product in excess. Possible causes include if their promoter and/or enhancer has become mutated (e.g., the oncomouse: a transgenic mouse that has both copies of its myc gene under the influence of extra-powerful promoters) or loss (e.g., by a translocation) of the 3'-UTR of their mRNA so that a microRNA (miRNA) that normally represses translation can no longer do so.
All these oncogenes act as dominants; if the cell has one normal gene (called a proto-oncogene) and one mutated gene (the oncogene) at a pair of loci, the abnormal product takes control. No single oncogene can, by itself, cause cancer. It can, however, increase the rate of mitosis of the cell in which it finds itself. Dividing cells are at increased risk of acquiring mutations, so a clone of actively dividing cells can yield subclones of cells with a second, third, etc. oncogene. When a clone loses all control over its mitosis, it is well on its way to developing into a cancer.
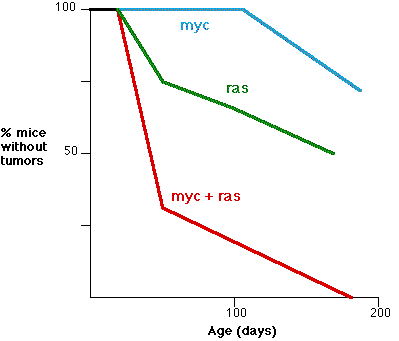
This graph (based on the work of E. Sinn et al, Cell 49:465,1987) shows the synergistic effect of two oncogenes. The fraction (%) of transgenic mice without tumors is shown as a function of age.
Other types of potential cancer-promoting genes
- Genes that inhibit apoptosis: The suicide of damaged cells — apoptosis — provides an important mechanism for ridding the body of cells that could go on to form a cancer. It is not surprising then that inhibiting apoptosis can promote the formation of a cancer. Example: Bcl-2. The product of this gene inhibits apoptosis. Overexpression of the gene is a hallmark of B-cell cancers.
- Genes involved in repairing DNA or stopping mitosis if they fail: Mutations arise from an unrepaired error in DNA. So any gene whose product participates in DNA repair probably can also behave as an oncogene when mutated. For example: ATM. ATM (="ataxia telangiectasia mutated") gets its name from a human disease of that name, whose patients — among other things — are at increased risk of cancer. The ATM protein is also involved in detecting DNA damage and interrupting the cell cycle when damage is found. It is estimated that fully 1% of the ~21,000 genes in the human genome are proto-oncogenes.
- Tumor-Suppressor Genes: The products of some genes inhibit mitosis. These genes are called tumor suppressor genes. In contrast to oncogenes, these behave as recessives — both alleles must be defective to lose their braking effect on mitosis.
Contributors and Attributions
John W. Kimball. This content is distributed under a Creative Commons Attribution 3.0 Unported (CC BY 3.0) license and made possible by funding from The Saylor Foundation.


