3.15: Junctions between Cells
- Page ID
- 3993
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)In many animal tissues (e.g., connective tissue), each cell is separated from the next by an extracellular coating or matrix. However, in some tissues (e.g., epithelia), the plasma membranes of adjacent cells are pressed together. Four kinds of junctions occur in vertebrates:
- Tight junctions
- Adherens junctions
- Gap junctions
- Desmosomes
In many plant tissues, it turns out that the plasma membrane of each cell is continuous with that of the adjacent cells. The membranes contact each other through openings in the cell wall called Plasmodesmata.
Tight Junctions
Epithelia are sheets of cells that provide the interface between masses of cells and a cavity or space (a lumen). The portion of the cell exposed to the lumen is called its apical surface. The rest of the cell (i.e., its sides and base) make up the basolateral surface. Tight junctions seal adjacent epithelial cells in a narrow band just beneath their apical surface. They consist of a network of claudins and other proteins. Tight junctions perform two vital functions:
- They limit the passage of molecules and ions through the space between cells. So most materials must actually enter the cells (by diffusion or active transport) in order to pass through the tissue. This pathway provides tighter control over what substances are allowed through.
- They block the movement of integral membrane proteins (red and green ovals) between the apical and basolateral surfaces of the cell. Thus the special functions of each surface, for example receptor-mediated endocytosis at the apical surface and exocytosis at the basolateral surface can be preserved.
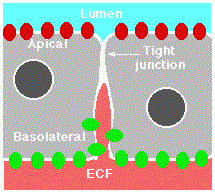
A report by Vermeer, et al., in the 20 March 2003 issue of Nature provides a striking example of the role of tight junctions. The epithelial cells of the human lung express
- a growth stimulant, called heregulin, on their apical surface and
- its receptors on the basolateral surface. (These receptors also respond to epidermal growth factor (EGF), and mutant versions have been implicated in cancer.
As long as the sheet of cells is intact, there is no stimulation of its receptors by heregulin thanks to the seal provided by tight junctions. However, if the sheet of cells becomes broken, heregulin can reach its receptors. The result is an autocrine stimulation of mitosis leading to healing of the wound. Several disorders of the lung the chronic bronchitis of cigarette smokers, asthma, cystic fibrosis increase the permeability of the airway epithelium. The resulting opportunity for autocrine stimulation may account for the proliferation (piling up) of the epithelial cells characteristic of these disorders.
Adherens Junctions
Adherens junctions provide strong mechanical attachments between adjacent cells. They hold cardiac muscle cells tightly together as the heart expands and contracts and hold epithelial cells together. Adherens junctions seem to be responsible for contact inhibition and some are present in narrow bands connecting adjacent cells. Others are present in discrete patches holding the cells together . Adherens junctions are built from cadherins that are transmembrane proteins (Figure 3.15.2; red labeled) whose extracellular segments bind to each other and whose intracellular segments bind to catenins (Figure 3.15.2; yellow labeled), which are connected to actin filaments
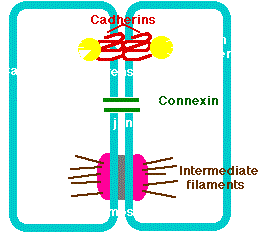
Human synthesize some 80 different types of cadherins. In most cases, a cell expressing one type of cadherin will only form adherens junctions with another cell expressing the same type. This is because molecules of cadherin tend to form homodimers, not heterodimers. Inherited mutations in a gene encoding a cadherin can cause stomach cancer. Mutations in a gene (APC), whose protein normally interacts with catenins, are a common cause of colon cancer. Loss of functioning adherens junctions may accelerate the edema associated with sepsis and tumor metastasis.
Gap Junctions
Gap junctions are intercellular channels some 1.5–2 nm in diameter. These permit the free passage between the cells of ions and small molecules (up to a molecular weight of about 1000 daltons). They are cylinders constructed from 6 copies of transmembrane proteins called connexins. Because ions can flow through them, gap junctions permit changes in membrane potential to pass from cell to cell.
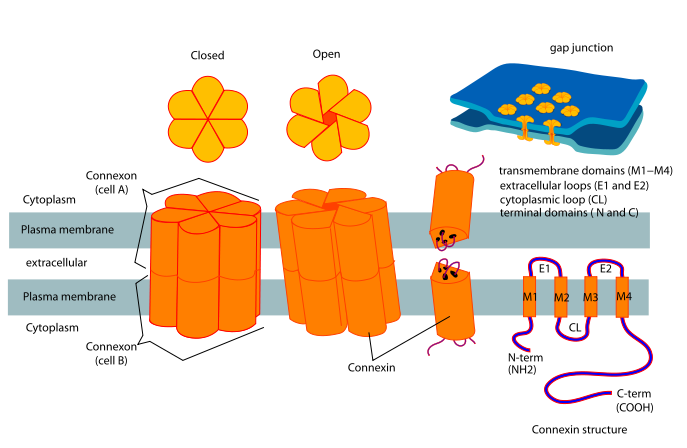
Examples of gap junctions include:
- The action potential in heart (cardiac) muscle flows from cell to cell through the heart providing the rhythmic contraction of the heartbeat.
- At some so-called electrical synapses in the brain, gap junctions permit the arrival of an action potential at the synaptic terminals to be transmitted across to the postsynaptic cell without the delay needed for release of a neurotransmitter.
- As the time of birth approaches, gap junctions between the smooth muscle cells of the uterus enable coordinated, powerful contractions to begin.
Several inherited disorders of humans such as certain congenital heart defects and certain cases of congenital deafness have been found to be caused by mutant genes encoding connexins.
Desmosomes
Desmosomes are localized patches that hold two cells tightly together. They are common in epithelia (e.g., the skin). Desmosomes are attached to intermediate filaments of keratin in the cytoplasm. Pemphigus is an autoimmune disease in which the patient has developed antibodies against proteins (cadherins) in desmosomes. The loosening of the adhesion between adjacent epithelial cells causes blistering. Carcinomas are cancers of epithelia. However, the cells of carcinomas no longer have desmosomes. This may partially account for their ability to metastasize.
Hemidesmosomes
These are similar to desmosomes but attach epithelial cells to the basal lamina ("basement membrane") instead of to each other. Pemphigoid is an autoimmune disease in which the patient develops antibodies against proteins (integrins) in hemidesmosomes. This, too, causes severe blistering of epithelia.
Plasmodesmata
Although each plant cell is encased in a boxlike cell wall, it turns out that communication between cells is just as easy, if not easier, than between animal cells (Figure 3.15.4). Fine strands of cytoplasm, called plasmodesmata, extend through pores in the cell wall connecting the cytoplasm of each cell with that of its neighbors.
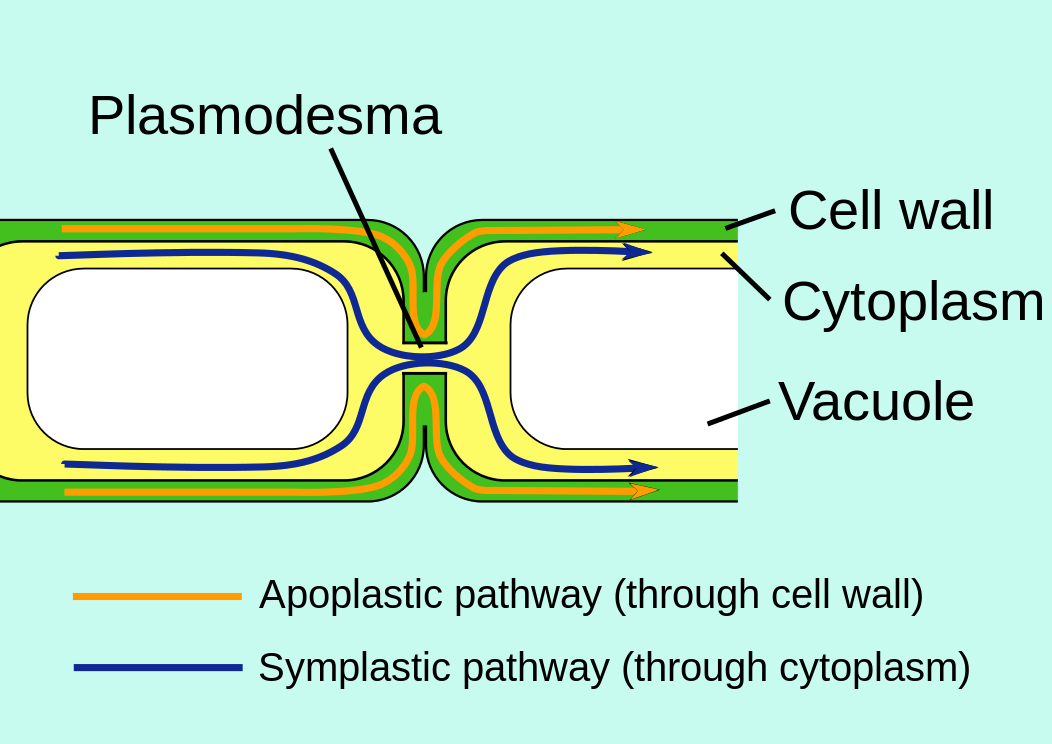
Plasmodesmata provide an easy route for the movement of ions, small molecules like sugars and amino acids, and even macromolecules like RNA and proteins, between cells. The larger molecules pass through with the aid of actin filaments. Plasmodesmata are sheathed by a plasma membrane that is simply an extension of the plasma membrane of the adjoining cells. This raises the intriguing question of whether a plant tissue is really made up of separate cells or is, instead, a syncytium: a single, multinucleated cell distributed throughout hundreds of tiny compartments.


