2.4: Polygenic inheritance and environmental effects
- Page ID
- 73733
How is height inherited?
If what you're really interested in is human genetics, learning about Mendelian genetics can sometimes be frustrating. You'll often hear a teacher use a human trait as an example in a genetics problem, but then say, "that's an oversimplification" or "it's much more complicated than that." So, what's actually going on with those interesting human traits, such as eye color, hair and skin color, height, and disease risk?
As an example, let's consider human height. Unlike a simple Mendelian characteristic, human height displays:
- Continuous variation. Unlike Mendel's pea plants, humans don’t come in two clear-cut “tall” and “short” varieties. In fact, they don't even come in four heights, or eight, or sixteen. Instead, it’s possible to get humans of many different heights, and height can vary in increments of inches or fractions of inches1.
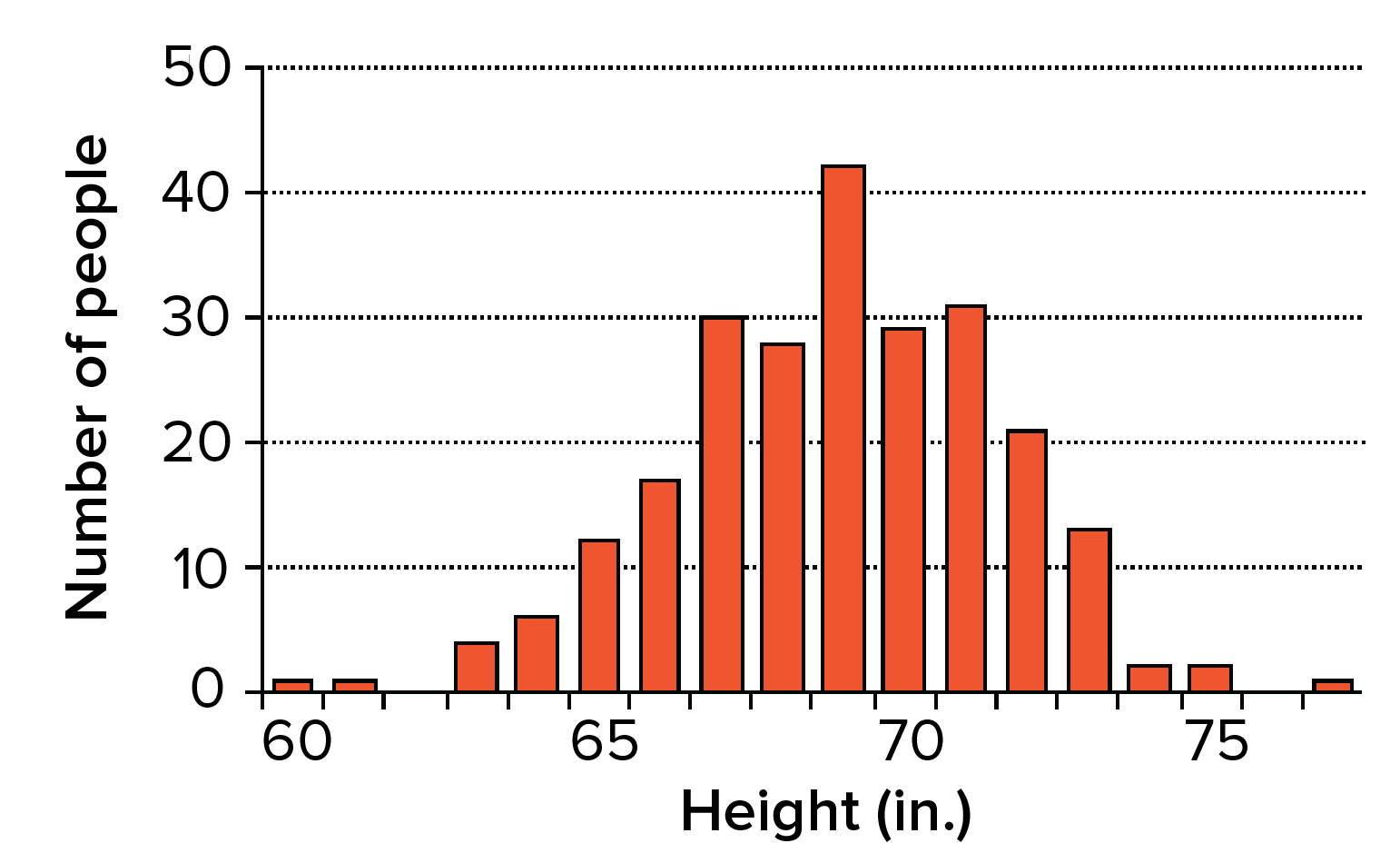
- A complex inheritance pattern. You may have noticed that tall parents can have a short child, short parents can have a tall child, and two parents of different heights may or may not have a child in the middle. Also, siblings with the same two parents may have a range of heights, ones that don't fall into distinct categories.
Simple models involving one or two genes can't accurately predict all of these inheritance patterns. How, then, is height inherited?
Height and other similar features are controlled not just by one gene, but rather, by multiple (often many) genes that each make a small contribution to the overall outcome. This inheritance pattern is sometimes called polygenic inheritance (poly- = many). For instance, a recent study found over 400 genes linked to variation in height2.
When there are large numbers of genes involved, it becomes hard to distinguish the effect of each individual gene, and even harder to see that gene variants (alleles) are inherited according to Mendelian rules. In an additional complication, height doesn’t just depend on genetics: it also depends on environmental factors, such as a child’s overall health and the type of nutrition he or she gets while growing up.
In this article, we’ll examine how complex traits such as height are inherited. We'll also see how factors like genetic background and environment can affect the phenotype (observable features) produced by a particular genotype (set of gene variants, or alleles).
Polygenic inheritance
Human features like height, eye color, and hair color come in lots of slightly different forms because they are controlled by many genes, each of which contributes some amount to the overall phenotype. For example, there are two major eye color genes, but at least 14 other genes that play roles in determining a person’s exact eye color3.
Looking at a real example of a human polygenic trait would get complicated, largely because we’d have to keep track of tens, or even hundreds, of different allele pairs (like the 400 involved in height!). However, we can use an example involving wheat kernels to see how several genes whose alleles "add up" to influence the same trait can produce a spectrum of phenotypes1,4.
In this example, there are three genes that make reddish pigment in wheat kernels, which we’ll call A, B, and C. Each comes in two alleles, one of which makes pigment (the capital-letter allele) and one of which does not (the lowercase allele). These alleles have additive effects: the aa genotype would contribute no pigment, the Aa genotype would contribute some amount of pigment, and the AA genotype would contribute more pigment (twice as much as Aa). The same would hold true for the B and C genes1,4.
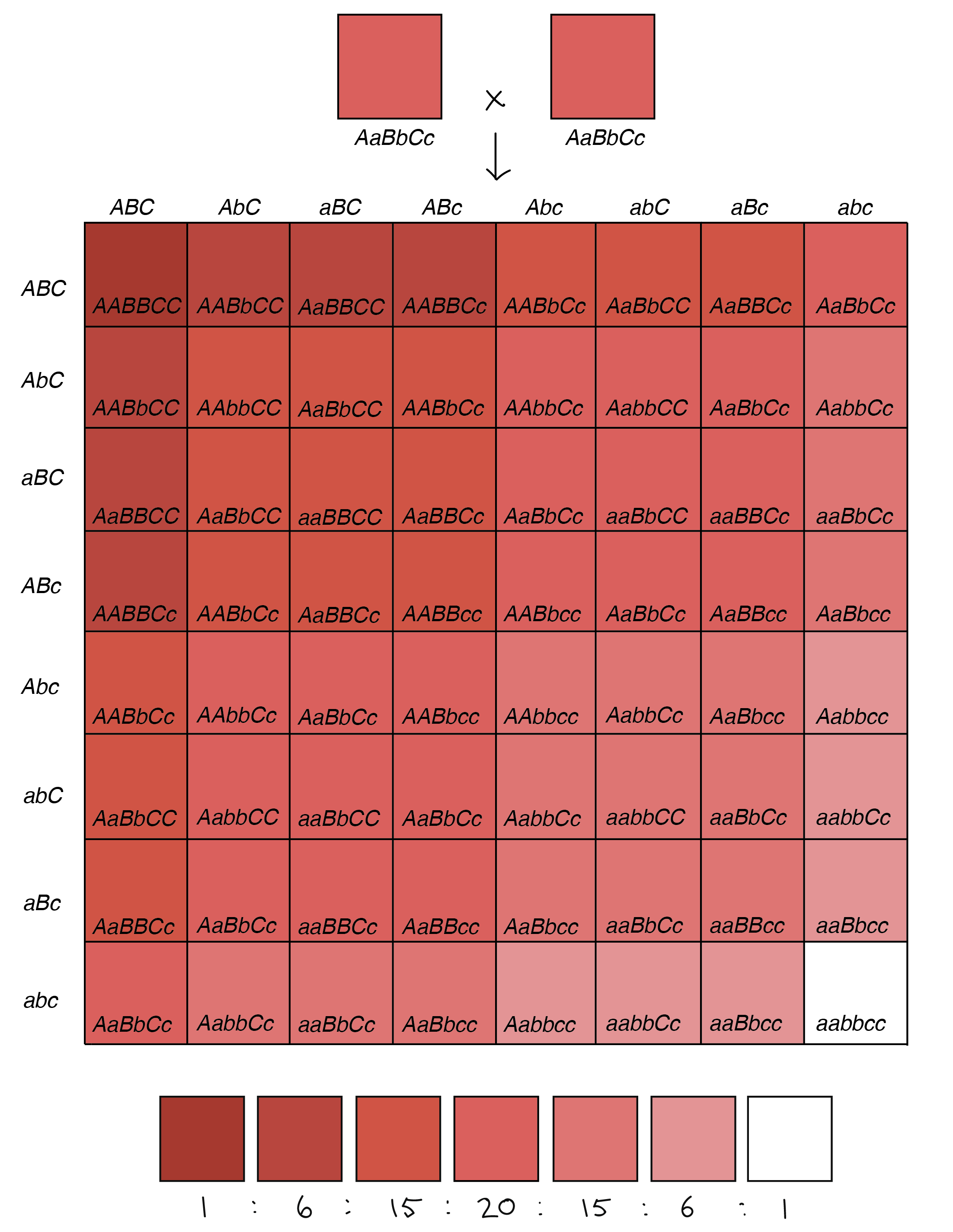
Now, let’s imagine that two plants heterozygous for all three genes (AaBbCc) were crossed to one another. Each of the parent plants would have three alleles that made pigment, leading to pinkish kernels. Their offspring, however, would fall into seven color groups, ranging from no pigment whatsoever (aabbcc) and white kernels to lots of pigment (AABBCC) and dark red kernels. This is in fact what researchers have seen when crossing certain varieties of wheat1,4.
This example shows how we can get a spectrum of slightly different phenotypes (something close to continuous variation) with just three genes. It’s not hard to imagine that, as we increased the number of genes involved, we’d be able to get even finer variations in color, or in another trait such as height.
Environmental effects
Human phenotypes—and phenotypes of other organisms—also vary because they are affected by the environment. For instance, a person may have a genetic tendency to be underweight or obese, but his or her actual weight will depend on diet and exercise (with these factors often playing a greater role than genes). In another example, your hair color may depend on your genes—until you dye your hair purple!
One striking example of how environment can affect phenotype comes from the hereditary disorder phenylketonuria (PKU)6. People who are homozygous for disease alleles of the PKU gene lack activity of an enzyme that breaks down the amino acid phenylalanine. Because people with this disorder cannot get rid of excess phenylalanine, it rapidly builds up to toxic levels in their bodies7.
If PKU is not treated, the extra phenylalanine can keep the brain from developing normally, leading to intellectual disability, seizures, and mood disorders. However, because PKU is caused by the buildup of too much phenylalanine, it can also be treated in a very simple way: by giving affected babies and children a diet low in phenylalanine8.
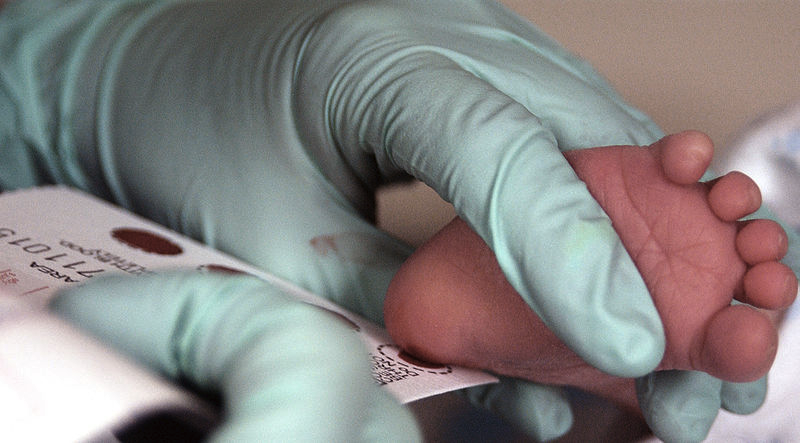
If people with phenylketonuria follow this diet strictly from a very young age, they can have few, or even no, symptoms of the disorder. In many countries, all newborns are screened for PKU and similar genetic diseases shortly after birth through a simple blood test, as shown in the image below.
Variable expressivity, incomplete penetrance
Even for characteristics that are controlled by a single gene, it’s possible for individuals with the same genotype to have different phenotypes. For example, in the case of a genetic disorder, people with the same disease genotype may have stronger or weaker forms of the disorder, and some may never develop the disorder at all.
In variable expressivity, a phenotype may be stronger or weaker in different people with the same genotype. For instance, in a group of people with a disease-causing genotype, some might develop a severe form of the disorder, while others might have a milder form. The idea of expressivity is illustrated in the diagram below, with the shade of green representing the strength of the phenotype.
- [Example]
-
The genetic disorder retinoblastoma causes cancerous tumors of the eyes, but the disease varies in severity and speed of onset. Children with retinoblastoma may develop tumors in just one eye or in both eyes, and the tumors may appear more quickly or slowly after birth9.

In incomplete penetrance, individuals with a certain genotype may or may not develop a phenotype associated with the genotype. For example, among people with the same disease-causing genotype for a hereditary disorder, some might never actually develop the disorder. The idea of penetrance is illustrated in the diagram below, with green or white color representing the presence or absence of a phenotype.
- [Example]
-
Some mutations in the retinoblastoma gene have high penetrance, but others have lower (incomplete) penetrance. For mutations in this latter category, some family members with the mutation are normal, while others develop retinoblastoma tumors6,11.
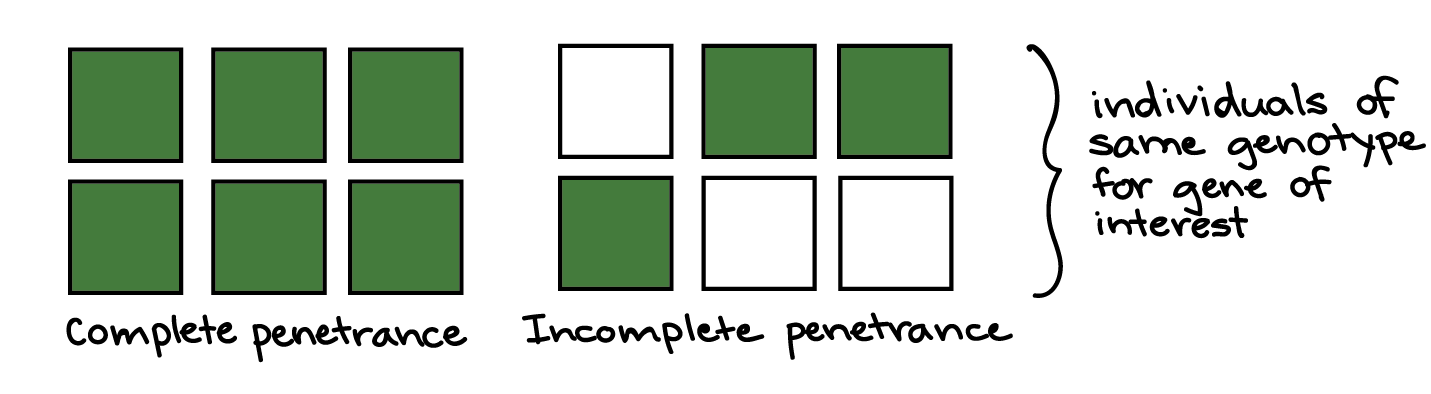
What causes variable expressivity and incomplete penetrance? Other genes and environmental effects are often part of the explanation. For example, disease-causing alleles of one gene may be suppressed by alleles of another gene elsewhere in the genome, or a person's overall health may influence the strength of a disease phenotype11.
Contributors and Attributions
Khan Academy (CC BY-NC-SA 3.0; All Khan Academy content is available for free at www.khanacademy.org)
- [Attribution and references]
-
This article is licensed under a CC BY-NC-SA 4.0 license.
Works cited:
-
Kimball, J. W. (2011, March 8). Continuous variation: Quantitative traits. Retrieved from http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/Q/QTL.html.
-
Wood, A. R., Esko, T., Yang, J., Vedantam, S., Pers, T. H., Gustafsson, S., ... Frayling, T. M. (2014). Defining the role of common variation in the genomic and biological architecture of adult human height. Nature Genetics, 46, 1173-1186. http://dx.doi.org/10.1038/ng.3097.
-
White, D. and Rabago-Smith, M. (2011). Genotype-phenotype associations and human eye color. Journal of Human Genetics, 56, 5-7. http://dx.doi.org/10.1038/jhg.2010.126.
-
Department of Agronomy, Iowa State University. (2016). Inheritance of a quantitative trait. Retrieved July 26, 2016 from https://masters.agron.iastate.edu/classes/527/lesson07/detail/kernelColor.html.
-
Armstrong, W. P. (n.d.). Continuous variation and Rh blood factor. In Wayne's Word. Retrieved July 26, 2016 from http://waynesword.palomar.edu/lmexer5.htm.
-
Bergmann, D. C. (2011). Variations on and complications of the basic Mendelian rules. In Genetics lecture notes. Biosci41, Stanford University, 10.
-
PAH. (2008). In Genetics home reference. Retrieved from http://ghr.nlm.nih.gov/gene/PAH.
-
Phenylketonuria. (2012). In Genetics home reference. Retrieved from http://ghr.nlm.nih.gov/condition/phenylketonuria.
-
National Cancer Institute. (2015, August 14). Retinoblastoma treatment – for health professionals. In Cancer types. Retrieved from http://www.cancer.gov/types/retinoblastoma/hp/retinoblastoma-treatment-pdq.
-
Carr, S. M. (2014). Penetrance versus expressivity. Retrieved from https://www.mun.ca/biology/scarr/Penetrance_vs_Expressivity.html.
-
Kelly, Jane. (2015, September 14). Retinoblastoma. In OMIM. Retrieved from http://www.omim.org/entry/180200.
-
Griffiths, A. J. F., Miller, J. H., Suzuki, D. T., Lewontin, R. C., and Gelbart, W. M. (2000). Penetrance and expressivity. In An introduction to genetic analysis (7th ed.). Retrieved from http://www.ncbi.nlm.nih.gov/books/NBK22090/.
References:
Armstrong, W. P. (n.d.). Continuous variation and Rh blood factor. In Wayne's Word. Retrieved July 26, 2016 from http://waynesword.palomar.edu/lmexer5.htm.
Bergmann, D. C. (2011). Variations on and complications of the basic Mendelian rules. In Genetics lecture notes. Biosci41, Stanford University, 8-12.
Breeding for grain quality traits: The challenges of measuring phenotypes and identifying genotypes. (2015). In Plant and soil sciences eLibrary. Retrieved from http://passel.unl.edu/pages/informationmodule.php?idinformationmodule=1066416033&topicorder=12&maxto=13.
Carr, S. M. (2014). Penetrance versus expressivity. Retrieved from https://www.mun.ca/biology/scarr/Penetrance_vs_Expressivity.html.
Department of Agronomy, Iowa State University. (2016). Inheritance of a quantitative trait. Retrieved July 26, 2016 from https://masters.agron.iastate.edu/classes/527/lesson07/detail/kernelColor.html.
Duffy, D. L., Montgomery, G. W., Chen, W., Zhao, Z. Z., Le, L., James, M. R., ... Sturm, R. A. (2007). A three–single-nucleotide polymorphism haplotype in intron 1 of OCA2 explains most human eye-color variation. American Journal of Human Genetics, 80(2), 241-252. http://dx.doi.org/10.1086/510885.
Griffiths, A. J. F., Miller, J. H., Suzuki, D. T., Lewontin, R. C., and Gelbart, W. M. (2000). Penetrance and expressivity. In An introduction to genetic analysis (7th ed.). Retrieved from http://www.ncbi.nlm.nih.gov/books/NBK22090/.
Kelly, Jane. (2015, September 14). Retinoblastoma. In OMIM. Retrieved from http://www.omim.org/entry/180200.
Multifactorial traits: Genomics at the cellular and gene level. (n.d.). In MI genetics resource center. Retrieved from https://www.migrc.org/TeachersAndStudents/MultifactorialTraits.html
National Cancer Institute. (2015, August 14). Retinoblastoma treatment – for health professionals. In Cancer types. Retrieved from http://www.cancer.gov/types/retinoblastoma/hp/retinoblastoma-treatment-pdq.
NHS National Genetics and Genomics Education Centre (n.d.). In Genetic glossary. Retrieved from http://www.geneticseducation.nhs.uk/genetic-glossary/221-penetrance.
Penetrance. (2015, November 13). Retrieved November 22, 2015 from Wikipedia: https://en.wikipedia.org/wiki/Penetrance.
PAH. (2008). In Genetics home reference. Retrieved from http://ghr.nlm.nih.gov/gene/PAH.
Phenylketonuria. (2012). In Genetics home reference. Retrieved from http://ghr.nlm.nih.gov/condition/phenylketonuria.
Polygenic. (2015, November 18). In Genetics home reference. Retrieved from http://ghr.nlm.nih.gov/glossary=polygenic.
Polygenic inheritance. (n.d.). In BioNinja. Retrieved from www.vce.bioninja.com.au/aos-3-heredity/inheritance/polygenic-inheritance.html.
Reece, J. B., Urry, L. A., Cain, M. L., Wasserman, S. A., Minorsky, P. V., and Jackson, R. B. (2011). Mendel and the gene idea. In Campbell biology (10th ed., pp. 267-291). San Francisco, CA: Pearson.
What are reduced penetrance and variable expressivity? (2015, November 18). In Genetics home reference. Retrieved from http://ghr.nlm.nih.gov/handbook/inheritance/penetranceexpressivity.
White, D. and Rabago-Smith, M. (2011). Genotype-phenotype associations and human eye color. Journal of Human Genetics, 56, 5-7. http://dx.doi.org/10.1038/jhg.2010.126.
Wood, A. R., Esko, T., Yang, J., Vedantam, S., Pers, T. H., Gustafsson, S., ... Frayling, T. M. (2014). Defining the role of common variation in the genomic and biological architecture of adult human height. Nature Genetics, 46, 1173-1186. http://dx.doi.org/10.1038/ng.3097.
Zeratsky, K. (2015, October 6). My favorite diet soda has a warning about phenylalanine. Is phenylalanine bad for you? [answer]. In Nutrition and healthy eating. Retrieved from http://www.mayoclinic.org/healthy-lifestyle/nutrition-and-healthy-eating/expert-answers/phenylalanine/faq-20058361.
Zlotogora, J. (2003). Penetrance and expressivity in the molecular age. Genetics in Medicine, 5, 347-352. http://dx.doi.org/10.1097/01.GIM.0000086478.87623.69.
-

