8.4: Propagating DNA in Bacteria
- Page ID
- 24782
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Complementation
George Beadle and Edward Tatum first described the concept that each gene corresponded to an enzyme in a metabolic pathway by exposing the yeast Neurospora crassa to mutagenic conditions (Beadle & Tatum, 1941). Following these procedures, Joshua Lederberg continued these studies with Tatum where they generated two mutants strains in Escherichia coli. These bacteria were auxotrophs, unable to generate some basic nutrients necessary to sustain their growth. The two strains were described as met− bio− Thr+ Leu+ Thi+ (Strain A) and Met+ Bio+ thr− leu− thi− (Strain B). Strain A can sufficiently synthesize the amino acids threonine, leucine, and the cofactor thiamine while deficient in producing the cofactor biotin and the amino acid methionine while the converse was true of Strain B. When either of these two strains was plated onto minimal media, no growth occurred. Supplementing minimal media with methionine and biotin permitted Strain A to grow as normal. When the two strains were mixed together and plated on minimal media, there was a growth of bacteria. The two strains were capable of complementing each other in some way as if a sexual exchange of genetic material had occurred (Lederberg & Tatum, 1946).
Bacteria are equipped with all the necessary capacities to replicate DNA. Common bacterial species have bee adapted for use in the lab to carry DNA and propagate it for uses in biotechnology. In addition to chromosomal DNA of the bacterial genome, bacteria also have extrachromosomal DNA called plasmids. These plasmids replicate independently of the bacterial chromosome and can occur in a high copy. These circular pieces of DNA are modified in labs to carry specific pieces of DNA so they can be studied or used for expression into proteins. Plasmids can naturally carry important traits, including antibiotic resistance. Plasmids are relatively small, ranging in size from 1000 bases to 1,000,000 bases long (1kb-1000kb).

Bacterial DNA usually exists as a large circular chromosome (red). Plasmids are extrachromosomal and autonomously replicating pieces of DNA (blue).
Through a process called conjugation, bacteria can “sexually” transfer genetic material to another by passing plasmids through a structure called a conjugation pilus.

Conjugation process between a plasmid bearing donor and a plasmid-less recipient. The donor creates a conjugation pilus to create a cytosolic bridge with the donor where the plasmid is replicated into the recipient through the rolling circle method of replication. The recipient then becomes competent to act as a donor.
Features of Plasmids
Plasmids that are designed by Biologists to shuttle pieces of DNA for study are referred to as vectors because they move a piece of DNA.
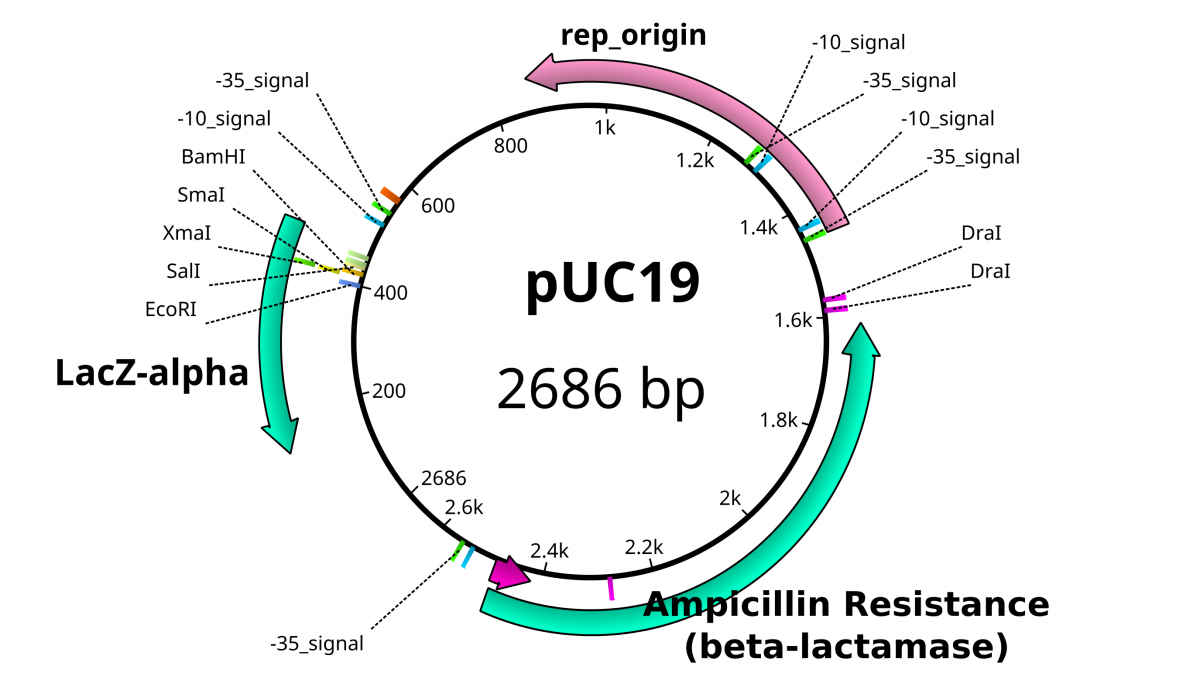
These plasmid vectors have the same hallmarks as traditional plasmids with the capacity to replicate independently of the bacterial genome. The feature that allows these DNA’s to replicate is called an origin of replication (ori) that is usually rich in A’s and T’s. However, these plasmid vectors have the additional properties that make them easy to work with and distinguishable from bacterial plasmids; a selection marker and a multiple cloning site. A selection marker usually comes in the form of a gene that encodes resistance to a specific antibiotic. In the pictured plasmid, Ampicillin resistance granted by the β-lactamase gene. The multiple cloning site (MCS), also known as the polylinker, is the location in which the DNA of interest is incorporated into the vector. MCSs are defined by a set of unique sites where the DNA can be cut by restriction endonucleases (RE). As the name implies, restriction enzymes are “restricted” in their ability to cut or digest DNA. The restriction that is useful to biologists is usually palindromic DNA sequences. Palindromic sequences are the same sequence forwards and backward. Some examples of palindromes: RACE CAR, CIVIC, A MAN A PLAN A CANAL PANAMA. With respect to DNA, there are 2 strands that run antiparallel to each other. Therefore, the reverse complement of one strand is identical to the other.


EcoRI generates sticky of cohesive ends SmaI generates blunt ends
Restriction enzymes hydrolyze covalent phosphodiester bonds of the DNA to leave either “sticky/cohesive” ends or “blunt” ends. This distinction in cutting is important because an EcoRI sticky end can be used to match up a piece of DNA cut with the same enzyme in order to glue or ligate them back together. While endonucleases cut DNA, ligases join them back together. DNA digested with EcoRI can be ligated back together with another piece of DNA digested with EcoRI, but not to a piece digested with SmaI. Another blunt cutter is EcoRV with a recognition sequence of GAT | ATC.
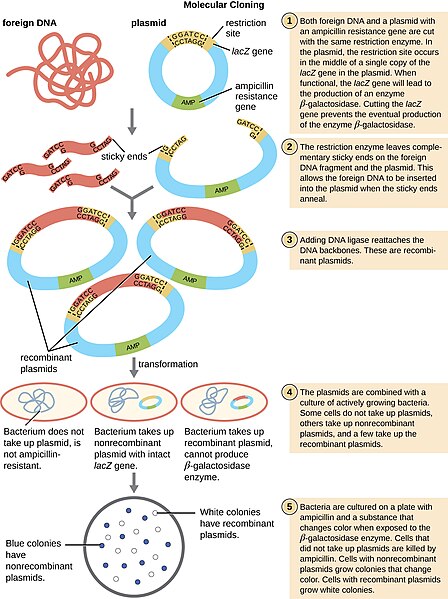
By “cutting and pasting” DNA into vectors, we can introduce foreign or exogenous DNA into bacteria. This type of DNA is now called Recombinant DNA and is the heart of biotechnology.
Recombinant DNA Technology

Additional Resources
Questions for Thought
1. Why do you think that the origins of replication are made of A’s and T’s?
2. What is different about the types of bonds holding the double strands together versus phosphodiester bonds of the DNA backbone?
3. Can DNA be digested with SmaI be ligated to DNA digested with EcoRV?
4. If so, which enzyme will be able to digest this new DNA?
- Beadle, G. W.; Tatum, E. L. (1941). “Genetic Control of Biochemical Reactions in Neurospora”. Proceedings of the National Academy of Sciences. 27 (11): 499–506. doi:10.1073/pnas.27.11.499. PMC 1078370. PMID 16588492
- Lederberg J, Tatum EL (1946). “Gene recombination in E. coli“. Nature. 158 (4016): 558. doi:10.1038/158558a0


