8.6: DNA Fingerprinting (RFLP)
- Page ID
- 24775
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Restriction fragment length polymorphism (RFLP) is a technique that exploits variations in DNA sequences. DNA from differing sources will have variations or polymorphisms throughout the sequence. Using Restriction Enzymes, these differences in sequences may be teased out. However, if one were to take the entirety of the human genome and chop it up with a restriction enzyme, many indecipherable fragments would be made. In fact, the resulting agarose gel would simply show a large smear of DNA. RFLP analysis requires that a probe to a specific area of DNA be used to identify specific locations. Agarose gels would be transferred to a membrane or filter where they would be hybridized to these radioactive probes.
Homologous chromosomes with restriction sites noted by triangles. the rectangle sitting on the chromosomes correspond to a probe locus. Credit: Jeremy Seto (CC0)
RFLP analysis was designed for forensic science to discriminate between people. Since people are 2N, they have pairs of homologous chromosomes with the same loci. However, these loci may contain different alleles. In this case, the phenotype for these alleles is the actual sequence that may or may not contain restriction sites. The presence or absence of a restriction site may arise from single nucleotide polymorphisms (SNPs) that reveal the natural variation between people.
The schematic below illustrates a comparison of restriction profiles between two sources. Note that the probe overlaps a restriction site in one of the alleles. This probe will be able to bind to both fragments given sufficient sequence overlap. Upon resolving on an agarose gel, genomic DNA that does not hybridize with the probe will obscure the locus of interest as a large smear. A filter is placed on top of the agarose and pressed against it to transfer the DNA in a process called Southern Blotting. Following a lengthy transfer, the filter is denatured to and incubated with the radioactive probe. To visualize this probe hybridization, a film is exposed to the filter and processed.
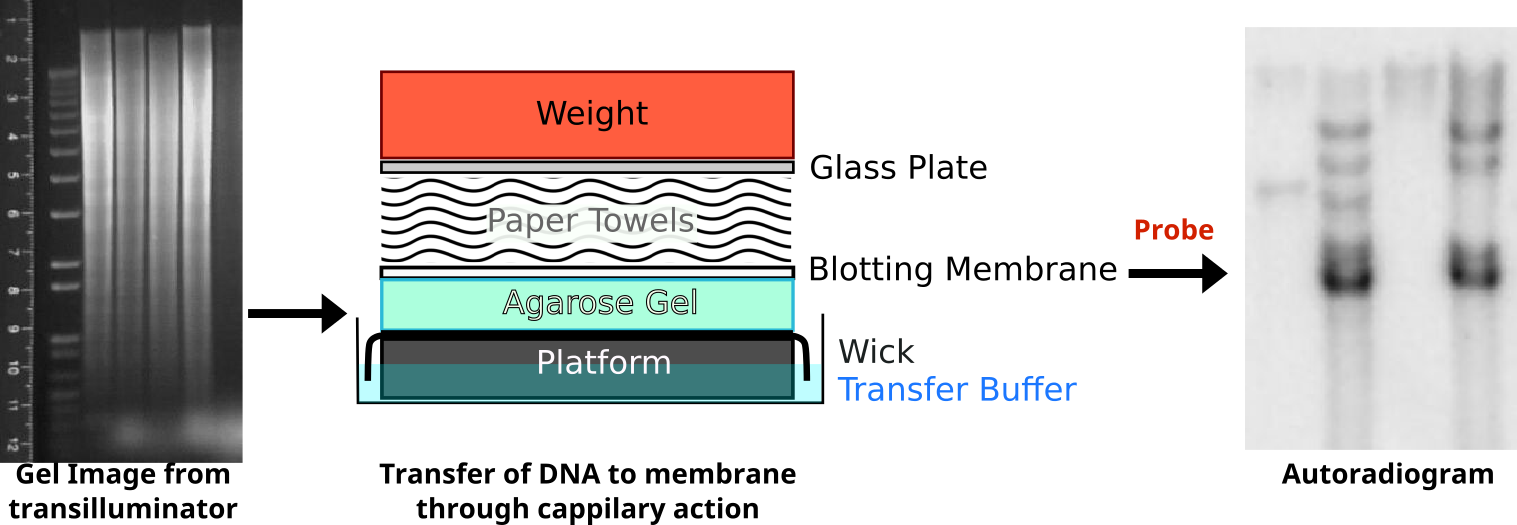
Following restriction digestion, the samples are resolved on an agarose gel. Digestion of genomic DNA will result in a large smear. Following transfer of the DNA onto a membrane through capillary action, the membrane is probed with radioactive probe DNA. The probe binds selectively to complementary sequences to reveal a series of distinct bands. An interactive demonstration of the first DNA fingerprinting.
Credit: Oder Zeichner: abigail [ or CC-BY-SA-3.0] /Autoradiogram
Sample A only reveals one band after processing because this person is homologous for the same allele. Sample B is heterozygous and reveals three bands.
Credit: Retama (CC-BY-SA 4.0)
RFLPs represent inheritable markers and can reveal relationships between different individuals. A pedigree can illustrate the relationship of the inherited alleles. The technique can be more informative if using multiple probes simultaneously for different loci or to use multi-locus probes that hybridize to multiple locations.
RFLPs may arise from differences in the STR/VNTR repeats between restriction sites. Credit: Jeremy Seto (CC0)
While RFLPs can arise from SNPs, they may also be caused by the expansion or contraction of repeated elements between restriction sites. These repeated elements of DNA are referred to as Variable Number Tandem Repeats (VNTR) and illustrate polymorphisms that normally occur in non-coding regions of the genome.
Related Pages
- Restriction Enzymes
- Analyzing DNA
- VNTR
- Genetics
External Resources
- Using Tandem Repeats for DNA Fingerprinting (Interview)
- The first DNA fingerprint (Flash Simulation)
DNA Fingerprinting (Activity)
- The origin of the DNA samples for this exercise will be explained by the Instructor as numerous scenarios may be used (Edvotek Cat. #109).
- Prepare a 1% agarose gel by adding 60ml Tris-Borate-EDTA buffer (TBE) to 0.6g agarose in an Erlenmeyer flask.
- Place the flask in a microwave or on the heat until agarose is melted.
- Stop periodically and swirl the solution. Do not permit to boil over.
- Assemble the casting tray by blocking the ends with tape or plastic baskets.
- Place the comb into the casting tray at the NEGATIVE end.
- The instructor will add 6μl Sybr Safe to his/her own gel solution at this time.
- You may place the casting trays inside a refrigerator and pour the solution into the tray.
- Wait until the gel is solidified.
- Carefully separate the gaskets from the tray ensuring not to tear apart the wells made by the comb.
- Remove the comb and place the casting tray into an electrophoresis chamber.
- Cover the gel with TBE buffer.
- Using a micropipettor, load 40-50μl dye samples sequentially into the wells.
- Cover the electrophoresis chamber with the lid and ensure good contact between electrodes.
- It is conventional that the POSITIVE side of the tank is nearest to you.
- With the POSITIVE side nearest to you, load the samples from left to right.
- Set the power supply to 100-120V, press the Run button (you should see bubbles at each electrode), and allow to run for at least 40 minutes.
- After 40 minutes, stop the current and remove the gel in the casting tray.
- Slide the gels into the staining solution if they do not include Sybr Safe for visualization the subsequent meeting time.
- The instructor will slide the gel onto a UV transilluminator behind a shield and show the results to the class.
- Document the findings of the gel by photographing with your phone.
- The instructor will discuss the results and ask for you to interpret the findings.
Activity Follow-up
1. Why are the samples loaded at the negative side of the gel?
2. What is the role of the dye in these samples? Should we be alarmed that the samples are all the same color?
3. What does it mean that there are multiple bands in a lane? What does it mean that there is only one band in a lane?
4. What can we conclude from the banding patterns in this forensics or paternity case? Is this sufficient data for these conclusions?


