2.4: Central Dogma and the Genetic Code
- Page ID
- 18129
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The "Central Dogma"
We have seen how DNA, with the aid of specific polymerases and accessory proteins, is able to replicate. We have also seen how we can use this information to create autonomously replicating extra-chromosomal elements (i.e. plasmids). However, the real utility of such systems arises when we use them to create proteins of interest. To get to proteins we have to go through RNA first.
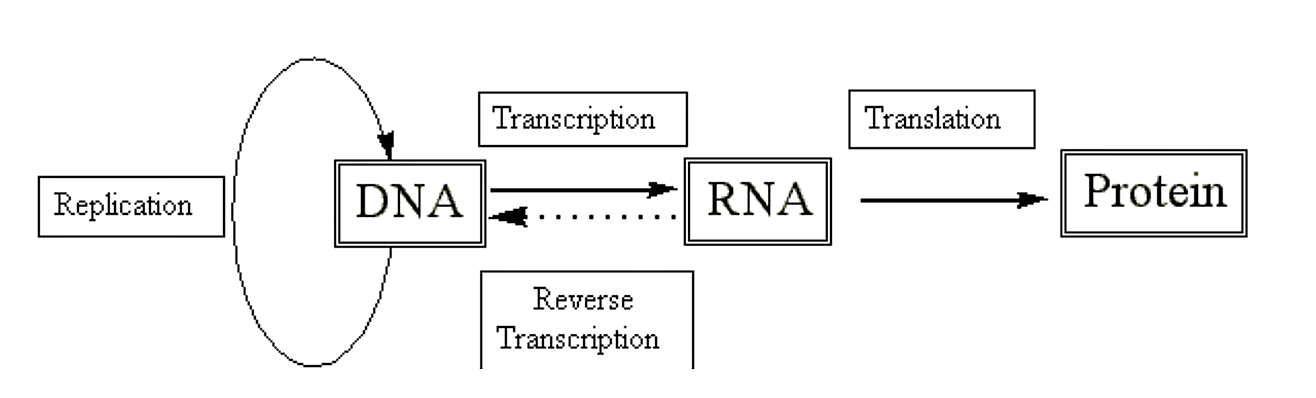.png?revision=1&size=bestfit&width=712&height=215)
Figure 2.4.1: Central dogma
Structural features of RNA:
- Similar to DNA except it contains a 2' hydoxyl group (makes phosphodiester bond more labile than DNA).
- Thymine in DNA is replaced by Uracil in RNA
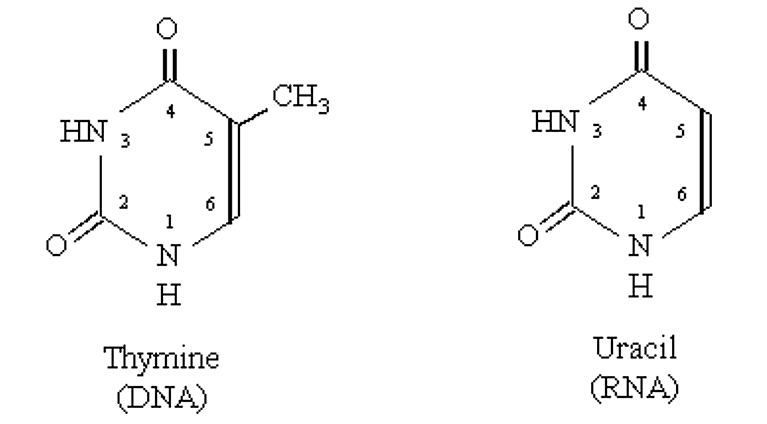.png?revision=1&size=bestfit&width=424&height=234)
Figure 2.4.2: Thymine vs. Uracil
3. RNA's can adopt regular three-dimensional structures which allow them to function in the process of genetic expression (i.e. the production of proteins).
- This ability to adopt defined three dimensional structures which impart functionality places RNA in a unique class - somewhat akin to proteins, and different from DNA.
- For example certain RNA molecules, when folded, exhibit catalytic capacities (e.g. the cleavage of RNA molecules).
- The majority of RNA in cells is found in complexes with proteins. The most common example is ribosomes (involved in protein synthesis).
Transcription: the copying of DNA by an RNA polymerase to make RNA.
RNA polymerase:
- Can initiate a new nucleic acid strand given a template.
- DNA polymerases cannot; they require a primer (or more typically, an RNA polymerase to provide the primer).
Protein Synthesis
- Three kinds of RNA molecules perform different functions in the protein synthesizing apparatus:
- Messenger RNA (mRNA) encodes the genetic information copied from DNA in the form of a sequence of bases that specifices a sequence of amino acids
- Transfer RNA (tRNA) is part of the structural machinery which deciphers the mRNA code. They carry specific amino acids which are transfered to a nacent polypeptide according to the instructions contained within the mRNA.
- Ribosomal RNA (rRNA) forms a complex with specific proteins to form the ribosome which is the key translational component
-
- the ribosome complexes with mRNA and directs appropriate tRNA's and the synthesis of the polypeptide bond.
Translation:
The process by which the information contained within a mRNA is used to direct the synthesis of the corresponding polypeptide.
The Genetic Code
How is the information for a polypeptide sequence stored within an mRNA molecule? There are twenty different common amino acids, but only four different bases in RNA (A, C, G, and U).
|
Base Arrangement |
Possible Combinations |
|---|---|
|
1 |
41=4 |
|
2 |
42=16 |
|
3 |
43=64 |
|
4 |
44=256 |
A triplet arrangement would seem to be the minimum possible combination necessary to code for the 20 different amino acids. Although, there are obviously going to be a lot of codons "left over". Most amino acids are coded for by more than a single unique triplet, and therefore the genetic code is said to be degenerate.
Experiments which led to the solution of the genetic code:
Nirenberg and Matthei (1961): Nirenberg and Matthei worked with bacterial extracts which contained everything needed for translation, with the exception of mRNA. To this they added either poly A, poly U or poly C RNA. The proteins produced by the translation of these RNA's was determined (poly G did not work, probably due to conformational problems):
|
Poly U |
Poly A |
Poly C |
|---|---|---|
|
Phe |
Lys |
Pro |
Thus, the triplet UUU = Phe, AAA = Lys, and CCC = Pro.
Korana (1963): In a cell free extract system, Korana added mRNA with repeating nucleotide sequences. The sequence ...ACACACAC... resulted in a polypeptide with alternating threonine and histidine residues. But, was threonine coded by ACA, and histidine by CAC? Or vise versa? To determine the answer to this, the mRNA sequence ...AACAACAACAAC... was tried. There were three different possible reading frames for the translation of this mRNA:
- AAC AAC AAC
- ACA ACA ACA
- CAA CAA CAA
But CAC was not a possible triplet. This sequence was found to code for three different polypeptide chains: poly Asn, poly Thr, and poly Gln. Since no histidine was found, histidine was therefore coded for by the triplet CAC.
Nirenberg and Leder (1964): Nirenberg and Leder used a filter which would allow RNA triplets and charged tRNA's to pass through, but would prevent passage of larger ribosomes. Specific triplet RNA sequences would bind to ribosomes and cause the binding of the associated charged tRNA molecules (coded for by the specific triplet). In a given experiment, if a unique charged tRNA were radiolabeled (on the amino acid), then it could be determined whether that particular charged tRNA was associated for by the unique triplet. In this way, all 61 codons for amino acids were determined.
.png?revision=1&size=bestfit&width=675&height=408)
Figure 2.4.3: Nirenberg and Leder experiment
The genetic code
|
5' End (Start) |
Second Position |
3' End |
|||
|---|---|---|---|---|---|
|
|
U |
C |
A |
G |
|
|
U |
Phe |
Ser |
Tyr |
Cys |
U |
|
|
Phe |
Ser |
Tyr |
Cys |
C |
|
|
Leu |
Ser |
Stop |
Stop |
A |
|
|
Leu |
Ser |
Stop |
Trp |
G |
|
C |
Leu |
Pro |
His |
Arg |
U |
|
|
Leu |
Pro |
His |
Arg |
C |
|
|
Leu |
Pro |
Gln |
Arg |
A |
|
|
Leu |
Pro |
Gln |
Arg |
G |
|
A |
Ile |
Thr |
Asn |
Ser |
U |
|
|
Ile |
Thr |
Asn |
Ser |
C |
|
|
Ile |
Thr |
Lys |
Arg |
A |
|
|
Met (start) |
Thr |
Lys |
Arg |
G |
|
G |
Val |
Ala |
Asp |
Gly |
U |
|
|
Val |
Ala |
Asp |
Gly |
C |
|
|
Val |
Ala |
Glu |
Gly |
A |
|
|
Val |
Ala |
Glu |
Gly |
G |
Note
E. coli codon preferences are indicated.
All proteins in prokaryotes and eukaryotes begin translation with the initiator codon AUG (methionine). The three codons, UAA, UGA and UAG are termination codons (don't code for any amino acids but signal the end of the protein chain).
Note the apparent relative importance of the middle base in the codon triplet
|
U |
C |
A |
G |
|---|---|---|---|
|
Phe Leu Ile Met Val |
Ser Pro Thr Ala |
Tyr His Gln Asn Lys Asp Glu Stop |
Cys Trp Arg Ser Gly Stop |
|
Hydrophobic |
Small/Polar |
Charged/Polar |
Polar |
Can common protein architectures be patterned by a simple quaternary pattern of residues?
The twenty common amino acids and their three-letter and single-letter acronyms:
|
Amino Acid |
Three letter acronym |
One letter acronym |
|---|---|---|
|
Alanine |
Ala |
A |
|
Cysteine |
Cys |
C |
|
Aspartic Acid |
Asp |
D |
|
Glutamic Acid |
Glu |
E |
|
Phenylalanine |
Phe |
F |
|
Glycine |
Gly |
G |
|
Histidine |
His |
H |
|
Isoleucine |
Ile |
I |
|
Lysine |
Lys |
K |
|
Leucine |
Leu |
L |
|
Methionine |
Met |
M |
|
Asparagine |
Asn |
N |
|
Proline |
Pro |
P |
|
Glutamine |
Gln |
Q |
|
Arginine |
Arg |
R |
|
Serine |
Ser |
S |
|
Threonine |
Thr |
T |
|
Valine |
Val |
V |
|
Tryptophan |
Trp |
W |
|
Tyrosine |
Tyr |
Y |


