13.3: Gluconeogenesis
- Page ID
- 15007
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Introduction
Gluconeogenesis is a metabolic pathway that results in the generation of glucose from non-carbohydrate carbon substrates such as lactate, glycerol, and glucogenic amino acids. It is one of the two main mechanisms humans and many other animals use to keep blood glucose levels from dropping too low (hypoglycemia). The other means of maintaining blood glucose levels is through the degradation of glycogen (glycogenolysis). Gluconeogenesis is a ubiquitous process, present in plants, animals, fungi, bacteria, and other microorganisms. In animals, gluconeogenesis takes place mainly in the liver and, to a lesser extent, in the cortex of the kidneys. This process occurs during periods of fasting, starvation, low-carbohydrate diets, or intense exercise and is highly endergonic. For example, the pathway leading from phosphoenolpyruvate to glucose-6-phosphate requires 6 molecules of ATP. Gluconeogenesis is often associated with ketosis. Gluconeogenesis is also a target of therapy for type II diabetes, such as metformin, which inhibits glucose formation and stimulates glucose uptake by cells.
Lactate is transported back to the liver where it is converted into pyruvate by the Cori cycle using the enzyme lactate dehydrogenase. Pyruvate, the first designated substrate of the gluconeogenic pathway, can then be used to generate glucose (Figure \(\PageIndex{1}\)). All citric acid cycle intermediates, through conversion to oxaloacetate, amino acids other than lysine or leucine, and glycerol can also function as substrates for gluconeogenesis. Transamination or deamination of amino acids facilitates the entry of their carbon skeleton into the cycle directly (as pyruvate or oxaloacetate), or indirectly via the citric acid cycle. Glycerol, which is a part of the triacylglycerol molecule, can be used in gluconeogenesis.
Gluconeogenesis is a pathway consisting of eleven enzyme-catalyzed reactions. The pathway can begin in the mitochondria or cytoplasm, depending on the substrate being used. Many of the reactions are the reversible steps found in glycolysis (Figure \(\PageIndex{1}\)). In humans, gluconeogenesis is restricted to the liver and a lesser extent the kidney.
Figure\(\PageIndex{1}\): Gluconeognesis
Bypass I (reverse of step 10 in glycolysis): Pyruvate to Phosphoenolpyruvate
The conversion of pyruvate into phosphoenolpyruvate requires two enzymatic steps and the formation of oxaloacetate as the intermediate. In all species, the formation of oxaloacetate from pyruvate and any other TCA cycle intermediates is restricted to the mitochondrion, and the enzymes that convert PEP to glucose are found in the cytosol. The location of the enzyme that links these two parts of gluconeogenesis by converting oxaloacetate to PEP, PEP carboxykinase, is variable by species: it can be found entirely within the mitochondria, entirely within the cytosol, or dispersed evenly between the two, as it is in humans. Transport of PEP across the mitochondrial membrane is accomplished by dedicated transport proteins; however, no such proteins exist for oxaloacetate. Therefore in species that lack intra-mitochondrial PEP, oxaloacetate must be converted into malate or aspartate, exported from the mitochondrion, and converted back into oxaloacetate to allow gluconeogenesis to continue.
Here is the net reaction: Pyruvate + ATP + HCO3- + GTP → PEP + ADP + GDP + CO2 ΔGo' = +0.2 kcal/mol (0.8 kJ/mol)
Figure\(\PageIndex{2}\) shows the overall reaction for the conversion of pyruvate to phosphoenol pyruvate.
Figure \(\PageIndex{2}\): Overall reaction for the conversion of pyruvate to phosphoenol pyruvate
In systems that produce oxaloacetate in the mitochondria and then need to transport it to the cytosol to be converted into phosphoenolpyruvate, three enzymes are needed for the process: pyruvate carboxylase (PC) located in the mitochondrial matrix, aspartate aminotransferase (AAT), located in the matrix and the cytosol, and the phosphoenolpyruvate carboxykinase (PCK) located in the cytosol. Oxaloacetate and aspartate are intermediate compounds formed in the process.
In the first reaction, pyruvate carboxylase converts pyruvate into oxaloacetate, based on the following reaction:
Pyruvate carboxylase: pyruvate + HCO3- + ATP → oxaloacetate + ADP + Pi
The pyruvate carboxylase enzyme requires biotin as a cofactor and has two major enzymatic functions: 1) carbon fixation from carbon dioxide, and 2) carbon transferase activity, placing the carbon dioxide that has been fixed, onto the molecule of pyruvate to create oxaloacetate. The biotin cofactor is shown in Figure (\PageIndex{3}\). Biotin is a water-soluble vitamin (known as D-biotin or vitamin B7) and is a cofactor in many enzymatic reactions, especially those involving carboxylation (or carbon fixation reactions). It is part of the vitamin B2 complex and is an essential vitamin for mammals. Deficiency results in dermatitis, loss of hair, and neurologic symptoms.
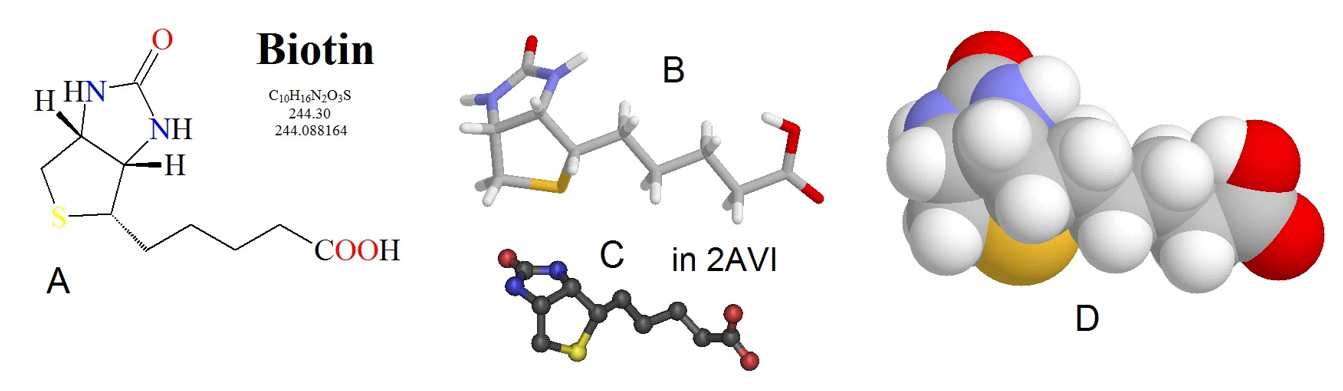
Figure (\PageIndex{3}\): Chemical Structure of Biotin. Biotin is shown in several different formats, (A) line structure, (B) stick model, (C) ball and stick model, and (D) spacefilling model.
Figure from: Biosynthesis
The biotin cofactor forms an amide linkage with the pyruvate carboxylase enzyme at a lysine residue (Figure (\PageIndex{4}\)). This creates a flexible linker region within the pyruvate carboxylase that is capable of dipping the biotin cofactor into the different catalytic domains of the enzyme to obtain the biological activity of the protein. Attaching the biotin to the enzyme requires the energy of ATP.
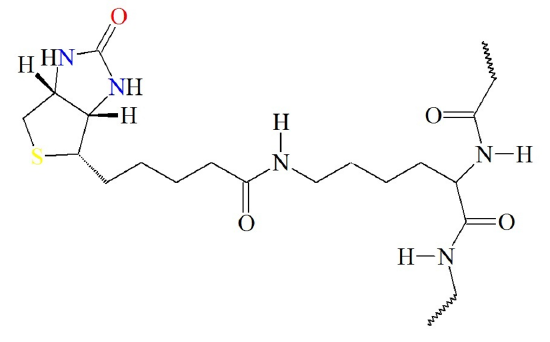
Figure (\PageIndex{4}\) Attachment of Biotin Cofactor to Lysine Residue in Pyruvate Carboxylase.
Image from: Biosynthesis
The pyruvate carboxylase enzyme is a tetramer that contains four functional protein subunits as shown in Figure \(\PageIndex{5}\). The BCCP flexible arm is capable of extending into the biotin carboxylation (BC) domain where biotin is first carboxylated, and then the BCCP shifts over to the carboxyl transferase (CT) domain where the fixed molecule of carbon dioxide can be transferred to pyruvate forming oxaloacetate.
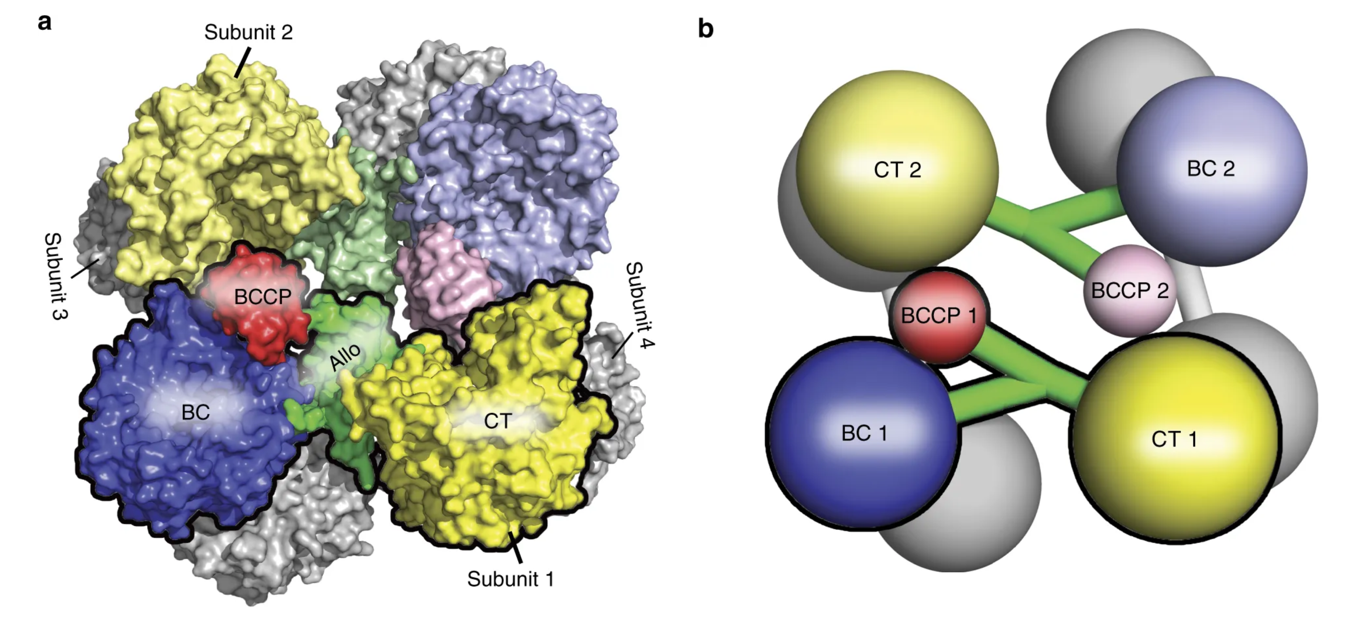
Figure \(\PageIndex{5}\) Structure of the Pyruvate Carboxylase Enzyme. The overall structure of the Pyruvate Carboxylase is a tetramer that contains four functional protein subunits. Two subunits are shown in color, with the other two indicated in gray at the back of the structure. (a) shows the space-filling model while (b) shows the major domains as a cartoon graphic. Each subunit contains a biotin carboxylase (BC) domain shown in blue, a carboxytransferase (CT) domain shown in yellow, and the biotin-carboxyl carrier protein (BCCP) domain shown in red and green.
Figure modified from Liu, Y., et al (2018) Nat Commun 9:1384
In the first part of the reaction, the biotin cofactor is carboxylated using bicarbonate as a substrate with ATP to drive the formation of a molecule with high energy with respect to its hydrolysis product intermediate. (Remember, there is no such thing as a "high energy" bond.) Thus, biotin is acting as an intermediate carrier of the carboxy group that will be added to pyruvate during the formation of oxaloacetate. This occurs in the biotin carboxylase domain (BC) with biotin bound to a biotin carboxyl carrier protein (BCCP) domain. The carboxyl group is then transferred to pyruvate to form oxaloacetate in the carboxyl transferase (CT) domain. Let's break up the mechanism of the enzyme from Rhizobium etli into three figures.
Figure\(\PageIndex{6}\) below shows the first steps in the carboxylation of biotin in the biotin carboxylase domain, using bicarbonate as a substrate. In Figure\(\PageIndex{6}\), you can see that an oxygen from bicarbonate mediates nucleophilic attack on the outer phosphate group of the ATP molecule forming the carbonyl-phosphate intermediate + ADP. The carbonyl-phosphate decomposes to release carbon dioxide and phosphate. The negatively charged phosphate activates the biotin cofactor to enable a nucleophilic attack on the carbon of the carbon dioxide producing the carboxybiotin intermediate, shown in the upper part of Figure\(\PageIndex{7}\).
Figure \(\PageIndex{6}\): The Carboxylation of Biotin during the Pyruvate Carboxylase Reaction Mechanism. Bicarbonate, shown in blue, enters the active site of the enzyme and reacts with the terminal phosphate of ATP forming a phosphorylated intermediate. This reaction is stabilized by two Mg2+ cofactors. The carboxyphosphate intermediate decomposes to release carbon dioxide and phosphate.
Image modified from Ribeiro AJM et al. (2017), Nucleic Acids Res, 46, D618-D623. Mechanism and Catalytic Site Atlas (M-CSA): a database of enzyme reaction mechanisms and active sites. DOI:10.1093/nar/gkx1012. PMID:29106569. https://www.ebi.ac.uk/thornton-srv/m-csa/entry/223/. Creative Commons Attribution 4.0 International (CC BY 4.0) License.
Figure\(\PageIndex{7}\): Carboxylation of biotin in pyruvate carboxylase. The upper diagram shows the actual carboxylation of biotin, which occurs in the BC domain. The carboxybiotin intermediate then shifts on the flexible arm and enters the carboxytransferase (CT) domain. Pyruvate enters the active site and this begins the steps involved in the carboxylation of pyruvate to form oxaloacetate. Ribeiro, ibid
The carboxyltransferase activity is housed in the CT domain of the enzyme and requires the formation of a carbanion intermediate to occur on the pyruvate molecule to make it reactive enough to cause the addition of the carboxyl group. A base from the enzyme eliminates a proton from the C3 position of pyruvate causing the formation of an enol reaction intermediate. As the electrons on the oxygen from the pyruvate molecule reform the carbonyl group, the electrons from the carbon-carbon double bond shift onto the carbon, creating a reactive carbanion intermediate. At this point, the carboxybiotinyl complex swings into the CT domain, providing the carboxyl group that will be incorporated into pyruvate. The carbanion intermediate mediates a nucleophilic attack onto the carboxyl carbon from the BCCP complex, and carbon dioxide is transferred to the pyruvate, creating oxaloacetate. The biotin cofactor is restored by the donation of the proton from the basic residue in the enzyme. This also restores the Pyruvate Carboxylase enzyme for another round of activity. Figure\(\PageIndex{8}\) below shows the carboxylation of the enolate form of pyruvate to form oxaloacetate.
Figure\(\PageIndex{8}\): Carboxylation of the enolate form of pyruvate to form oxaloacetate by pyruvate carboxylase. Ribeiro et al, ibid
Figure\(\PageIndex{9}\) below shows an interactive iCn3D model of the biotin-dependent multifunctional enzyme pyruvate carboxylase from Rhizobium etli (2QF7)
Figure\(\PageIndex{9}\): Biotin-dependent multifunctional enzyme pyruvate carboxylase from Rhizobium etli (2QF7). The enzyme is a homotetramer with C2 symmetry but only one subunit chain is shown for clarity. The biotin carboxylase domain (BC) is shown in cyan, the carboxytransferase domain (CT) in yellow, a C-terminal biotin carboxyl carrier protein (BCCP) domain (disconnected in parts) in magenta, and the allosteric domain in green. AGS in the cyan domain is a phosphothiophosphoric acid-adenylate ester, an ATP analog. It is shown interacting with the 2 Mg2+ ion and it is represented in sticks and labeled. Acetyl-CoA and a nonhydrolyzable analog, ethyl-CoA, are allosteric activators. CoASH (ethyl group not shown) is illustrated in spacefill bound to the green allosteric domain. Acetyl-CoA binding leads to a conformational change that decreases the distance between the two active sites on the BT (cyan) and CT (yellow) domains. Biotin, not shown, is tethered, in the BCCP domain, which transfers biotin between the two catalytic domains not within a single monomer (as shown above), but between different monomers in the homo 4-mer.
(Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...7g91JmzJMad4r9
Figure\(\PageIndex{10}\) shows the flexible nature of the biotin flexible arm of the enzyme during the catalytic process.
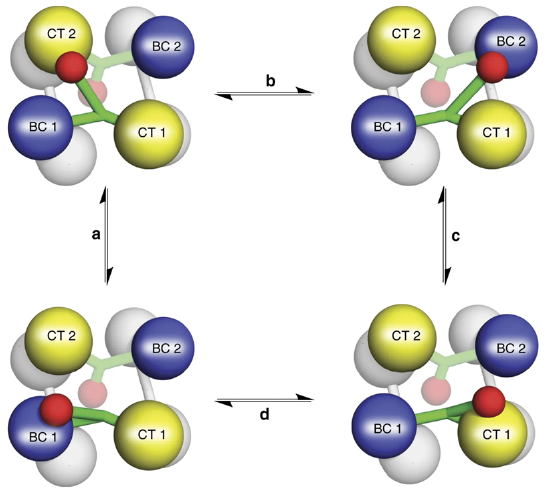
Figure\(\PageIndex{10}\) The Catalytic Activity of Pyruvate Carboxylase. The Biotin-Carboxyl Carrier Protein Domain (BCCP) is a flexible arm of the protein structure that can swing from one catalytic domain to the other during the reaction process. As shown in the diagram, the BCCP domain of subunit 1 will be fixed with carbon dioxide at the biotin carboxylase domain from subunit 1 (BC1). The BCCP will then translocate to the carboxyltransferase domain from subunit 2 (CT2) where it will create one molecule of oxaloacetate and reset the biotin cofactor. The BCCP will then swing into the BC2 domain where carbon fixation will occur. BCCP then shifts to the CT1 domain where it creates a second oxaloacetate molecule and resets the biotin. While this is going on with the BCCP from subunit 1, the other BCCP arms from subunits 2, 3, and 4 are also active. Thus, one full sequence of the enzyme activity will produce 8 molecules of oxaloacetate.
Image from: Liu, Y., et al (2018) Nat Commun 9:1384
The next reaction in the process requires the conversion of oxaloacetate to phosphoenolpyruvate by phosphoenolpyruvate carboxykinase (PEPCK). However, the major human PEPCK enzyme, PCK1, resides in the cytoplasm of the cell. Thus the oxaloacetate produced in the matrix of the mitochondria by pyruvate carboxylase must be transported into the cytosol. However, there are no oxaloacetate transporters that can mediate the transfer directly. To leave the mitochondrial matrix, oxaloacetate has to first be converted to aspartate by the aspartate aminotransferase enzyme (AAT). It can then be transported to the intermembrane space of the mitochondria through an antiporter that transports one molecule of aspartate out of the matrix and one molecule of glutamate into the matrix Figure\(\PageIndex{11}\). Once in the intermembrane space, the aspartate can pass freely through a pore in the outer mitochondrial membrane. When it reaches the cytoplasm, aspartate is reconverted to oxaloacetate using the cytoplasmic aspartate aminotransferase enzyme (AAT). Oxaloacetate can then be used as a substrate by the phosphoenolpyruvate carboxykinase enzyme (PEPCK).
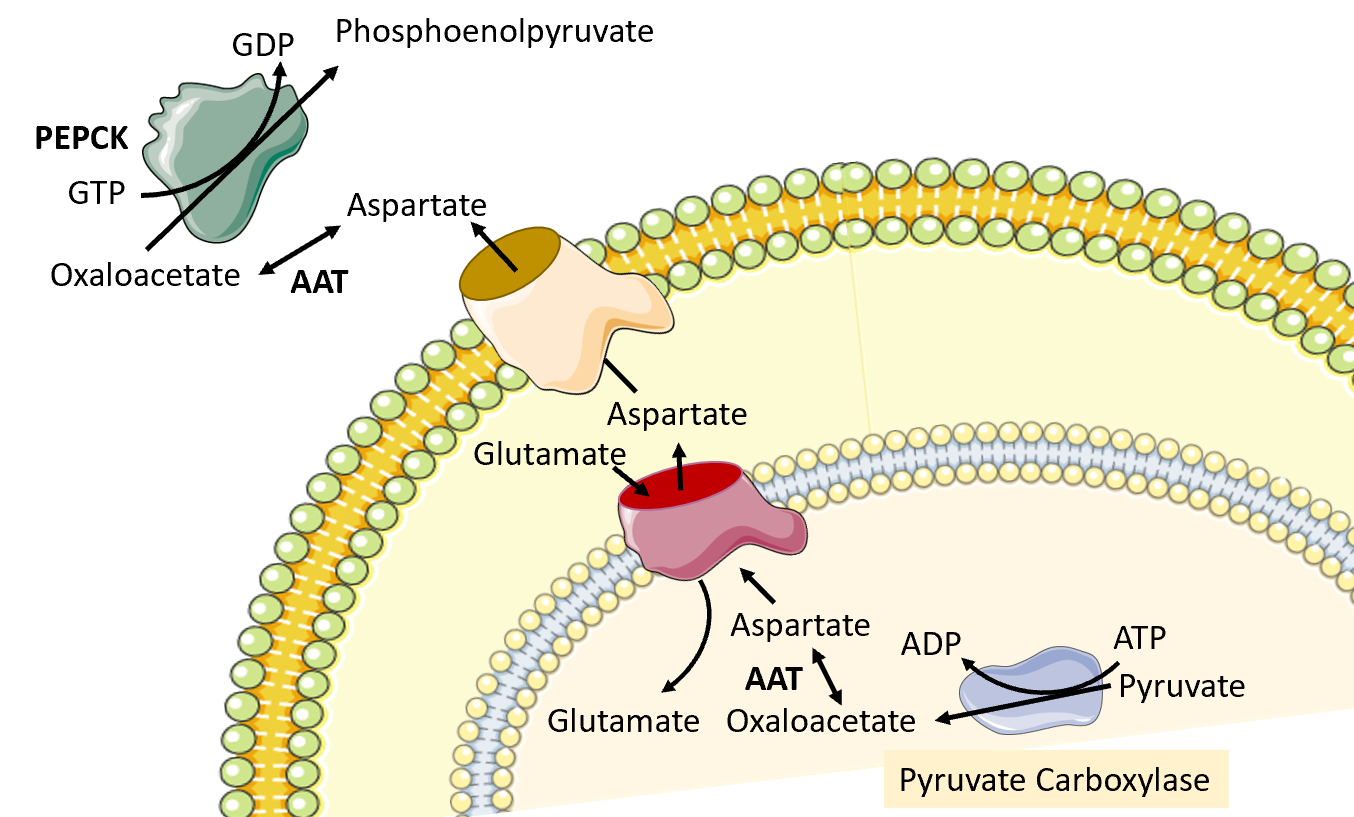
Figure\(\PageIndex{11}\) Conversion of Oxaloacetate to Aspartate and Transfer to the Cytoplasm.
Image modified from SMART Servier Medical Art
The glutamate/aspartate transporter has some additional complexities associated with it. It cannot function on its own. It requires the coordinated functioning of the malate/alpha-ketoglutarate antiporter. Together, these antiporters are known as the Malate-Aspartate Shuttle System.
The Malate-Aspartate Shuttle System is dependent on the functioning of two enzymatic processes. The first is the aspartate aminotransferase that was indicated more simplistically in Figure\(\PageIndex{11}\). The aspartate aminotransferase enzyme can utilize glutamate as an amine donor to generate aspartate from oxaloacetate. Alpha-ketoglutarate is also formed in this process. Depending on substrate concentrations and other regulatory mechanisms, this enzyme can also work in the reverse reaction to produce glutamate and oxaloacetate. In a different reaction using Malate Dehydrogenase, oxaloacetate can be reduced to form malate using a molecule of NADH as the electron donor.
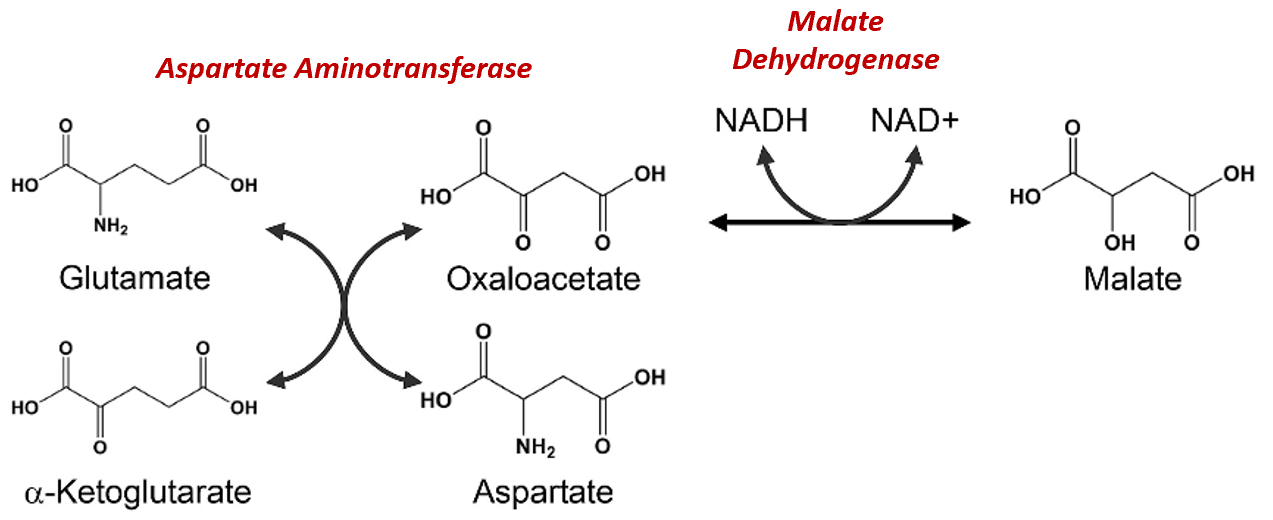
Figure\(\PageIndex{12}\) Chemical Reactions of the Malate-Aspartate Shuttle System
Figure modified from Son, H.F. and Kim, J-J. (2016) PLoS One 10.1371
The malate dehydrogenase enzyme is expressed to high levels in the cytoplasm of liver cells as well as in the matrix of the mitochondria. This enzyme is a component of the Kreb Cycle where it mediates the formation of oxaloacetate in the last step of the cycle. Within the cytoplasm, it predominantly converts oxaloacetate to malate. The malate can then be shuttled into the matrix of the mitochondria through the malate/alpha-ketoglutarate antiporter (Figure\(\PageIndex{13}\)). In this antiporter, malate moves into the mitochondrial matrix while alpha-ketoglutarate moves into the intermembrane space (and subsequently into the cytosol of the cell). Together with aspartate, it can be used by cytoplasmic aspartate aminotransferase to produce glutamate and oxaloacetate. This is the oxaloacetate pool that is then utilized in the gluconeogenic pathway when PEPCK is active. The glutamate generated from this reaction is transported back into the matrix of the mitochondria through the aspartate/glutamate antiporter. The pool of aspartate in the matrix of the mitochondria is supplied by the reaction of the aspartate aminotransferase enzyme, which completes the reverse reaction from the one seen in the cytoplasm. In the matrix, aspartate aminotransferase uses glutamate and oxaloacetate as substrates to generate aspartate and alpha-ketoglutarate. This enables the transport of aspartate and glutamate through their specific antiporter. Within the gluconeogenic pathway, heightened levels of oxaloacetate are produced in the matrix of the mitochondria. Oxaloacetate is then converted to aspartate and transported across the inner membrane, where it can subsequently be converted back into oxaloacetate and used for the production of glucose.
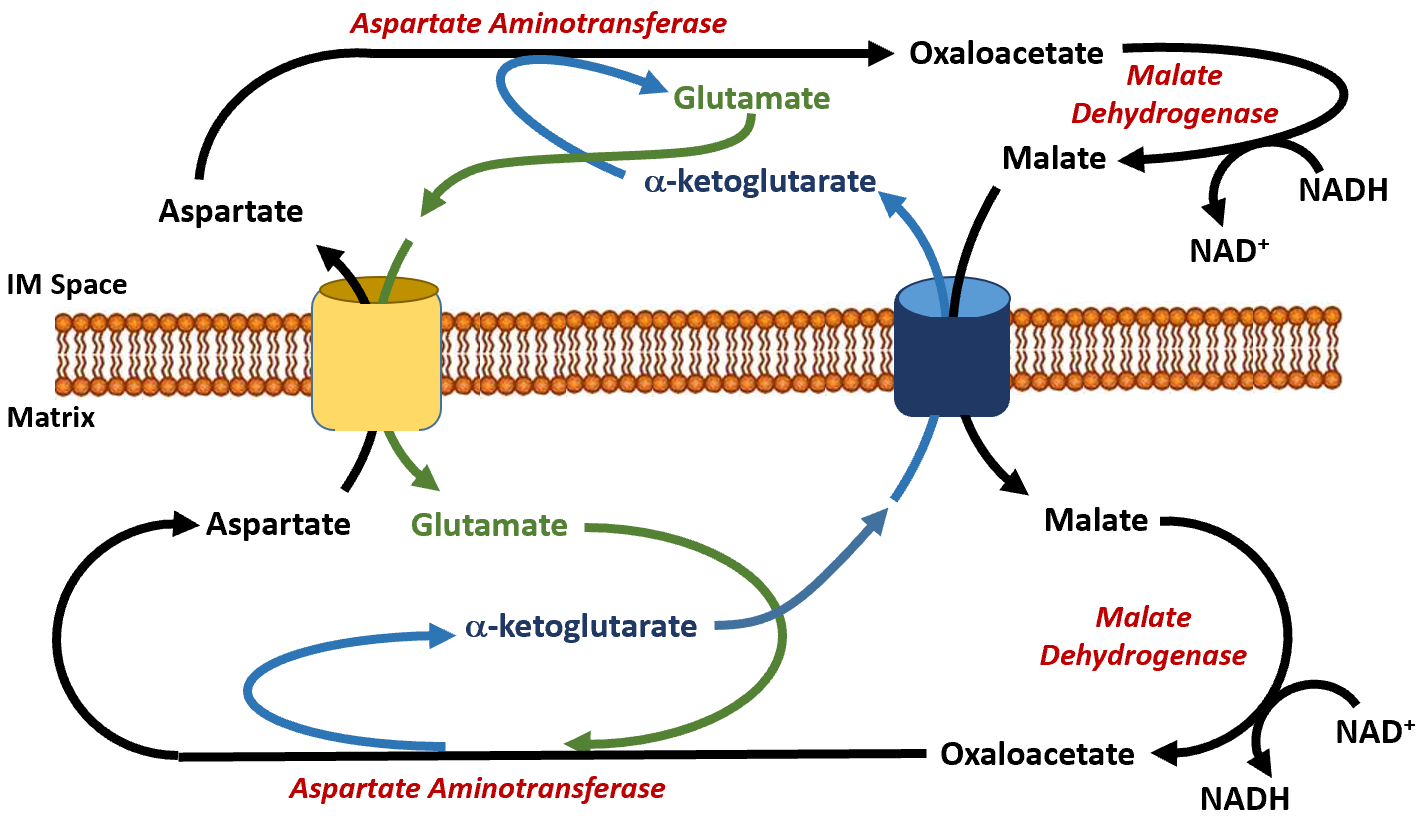
Figure\(\PageIndex{13}\): The Malate-Aspartate Shuttle System.
Once oxaloacetate has been effectively transported via the Malate-Aspartate Shuttle into the cytoplasm, it is converted to phosphoenolpyruvate (PEP) by the PEP carboxykinase (PEPCK). The overall reaction mediated by PEPCK is:
PEP carboxykinase (PEPCK): oxaloacetate + GTP → phosphoenol pyruvate + GDP
PEPCK is classified as a lyase enzyme and exists in two isozymes, a cytoplasmic PEPCK, and a mitochondrial PEPCK. The cytoplasmic form is the one that is predominantly used in the gluconeogenic pathway and requires the action of the Malate-Aspartate Shuttle. However, small amounts of PEP can be made directly by the mitochondrial PEPCK and then transported across the mitochondrial membrane. The conversion of oxaloacetate to phosphoenolpyruvate by PEPCK mediates the removal of carbon dioxide and the addition of a phosphate group. A molecule of GTP is used during this process as the phosphate donor and magnesium ions serve as a cofactor. Figure \(\PageIndex{14}\) shows the reaction mechanism for the human PEP carboxykinase.
Figure \(\PageIndex{14}\): Reaction mechanism for human PEP carboxykinase (PEPCK)
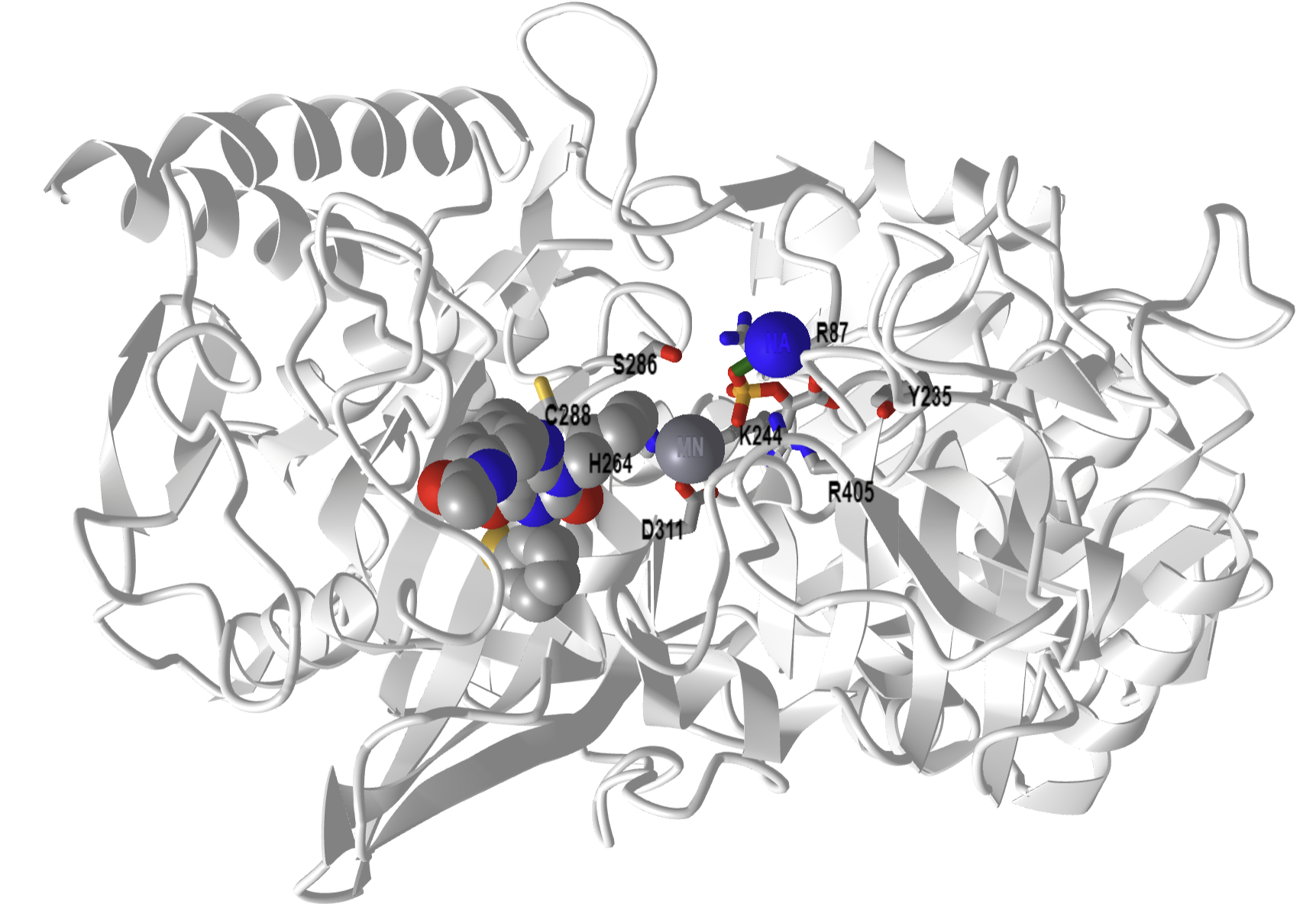
Figure\(\PageIndex{15}\) below shows an interactive iCn3D model of phosphoenolpyruvate carboxykinase (PEPK) with a bound PEP and GTP-competitive inhibitor (1NHX).
Figure \(\PageIndex{15}\): Phosphoenolpyruvate carboxykinase (PEPK) with a bound PEP and GTP-competitive inhibitor (1NHX). PEP is shown in colored sticks. The competitive inhibitor is shown in colored spacefill. The active site residues from the human PEPCK mechanism are shown as labeled sticks. (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...QFcTssCJbuW2K7
Phosphatase Reactions of Gluconeogenesis (reverse of steps 1 and 3 in glycolysis):
The last two unique enzymes of the gluconeogenic pathway are both phosphatase enzymes. Fructose 1,6-bisphosphatase converts fructose 1,6-bisphosphate to fructose 6-phosphate. The same phosphoglucoisomerase used in glycolysis can convert fructose 6-phosphate back into glucose 6-phosphate. A unique glucose 6-phosphatase enzyme will then convert glucose 6-phosphate to free glucose. The unique phosphatase reactions are shown in Figure\(\PageIndex{16}\).
Figure \(\PageIndex{16}\): Phosphorylase Enzymes utilized during Gluconeogenesis
First, let's consider the dephosphorylation of fructose 1,6-bisphosphate. The gluconeogenic enzyme is named fructose-1,6-bisphosphatase, (FBP or FBPase-1). FBP requires a metal cofactor and is competitively and allosterically regulated. Fructose 2,6-bisphosphate and low energy load (AMP and ADP) inhibit the enzyme. The overall reaction is shown below:
Fructose-1,6-bisphosphatase: F1,6BP + H2O → F6P + Pi
Figure \(\PageIndex{17}\) shows the mechanism of mouse fructose,1-6-bisphosphatase (FBPase-1)
Figure \(\PageIndex{17}\): Mechanism of mouse fructose,1-6-bisphosphatase. Mg2+ ions coordinate the positioning of F-1,6-BP near an activated water molecule that mediates nucleophilic attack on the 1-position phosphate group. This leads to the cleavage of the phosphate group from the sugar molecule, F6P, which can then leave the active site.
Figure\(\PageIndex{18}\) below shows an interactive iCn3D model of pig fructose-1,6-bisphosphatase with bound Mg2+, fructose-6-phosphate, and phosphate in the R state (1EYI).
Figure \(\PageIndex{18}\): Fructose-1,6-bisphosphatase with bound Mg, fructose-6-phosphate and phosphate in the R state (1EYI). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...DbWMHBRTdPGhJ8
FBP is an allosteric homotetramer and can exist in a tense (T) and relaxed (R) state. When the enzyme is in the R state, it has the highest catalytic activity. Figure\(\PageIndex{18}\) shows only one monomer of the R state with bound ligands and substrates for clarity.
Figure \(\PageIndex{19}\) shows an interactive iCn3D model of tetrameric FBP enzyme in the T state with bound Mg2+, fructose-6-phosphate, phosphate, and AMP, an allosteric inhibitor, in the T state (1EYJ).
Figure \(\PageIndex{19}\): Tetrameric fructose-1,6-bisphosphatase in the T state with bound Mg2+, fructose-6-phosphate, phosphate and AMP in the T state (1EYJ). (Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...UAbjNtkPsoBdA9
FBPase is inhibited by AMP and fructose-2,6-bisphosphate, and activated by ATP. Allosteric changes control the catalytic activity of the enzyme and the transition from the T to the R state, or vice versa. As shown in Figure\(\PageIndex{17}\), FBP has 3 metal binding sites. The mechanism shown in Figure \(\PageIndex{17}\) In the model, all of the binding sites are filled with Mg2+, however, Zn2+ can also play a role. In the presence of AMP, only one site is occupied by Mg2+, as shown in the model of the T state in Figure\(\PageIndex{19}\). The T state form of FBP has a loop (52-72) that is disordered. In the R state (no AMP), the loop interacts with the active site and 1 Zn2+ and 2 Mg2+ ions are bound in the three sites. The binding of AMP leads to the dissociation of two of the bound metal cofactors and causes a decrease in enzyme activity.
The resulting fructose 6-phosphate is isomerized to glucose 6-phosphate. Glucose 6-phosphate is then converted by the last unique gluconeogenic enzyme, glucose 6-phosphatase, to generate free glucose. The overall reaction is shown below:
Glucose-6-phosphatase: G6P + H2O → glucose + Pi
The dephosphorylation of glucose only occurs appreciably in liver cells, as this is the primary location for the regulation of blood glucose levels. This serves as the final step in the gluconeogenic pathway. The glucose 6-phosphatase enzyme is a transmembrane protein that resides in the inner membrane of the endoplasmic reticulum. Thus, for glucose 6-phosphate to be dephosphorylated, it must first be transported from the cytoplasm into the lumen of the endoplasmic reticulum (ER) through transporter 1 (T1) (Figure\(\PageIndex{20}\)). The glucose 6-phosphatase (G-6-Pase) then cleaves the phosphate from the substrate, releasing inorganic phosphate (P) and glucose (red molecule). The inorganic phosphate is then transported back into the cytoplasm through transporter 2 (T2) and glucose is transported through Transporter 3 (T3). Free glucose is then transported back into the bloodstream through a glucose (GLUT) transporter (not shown in Figure\(\PageIndex{20}\)).
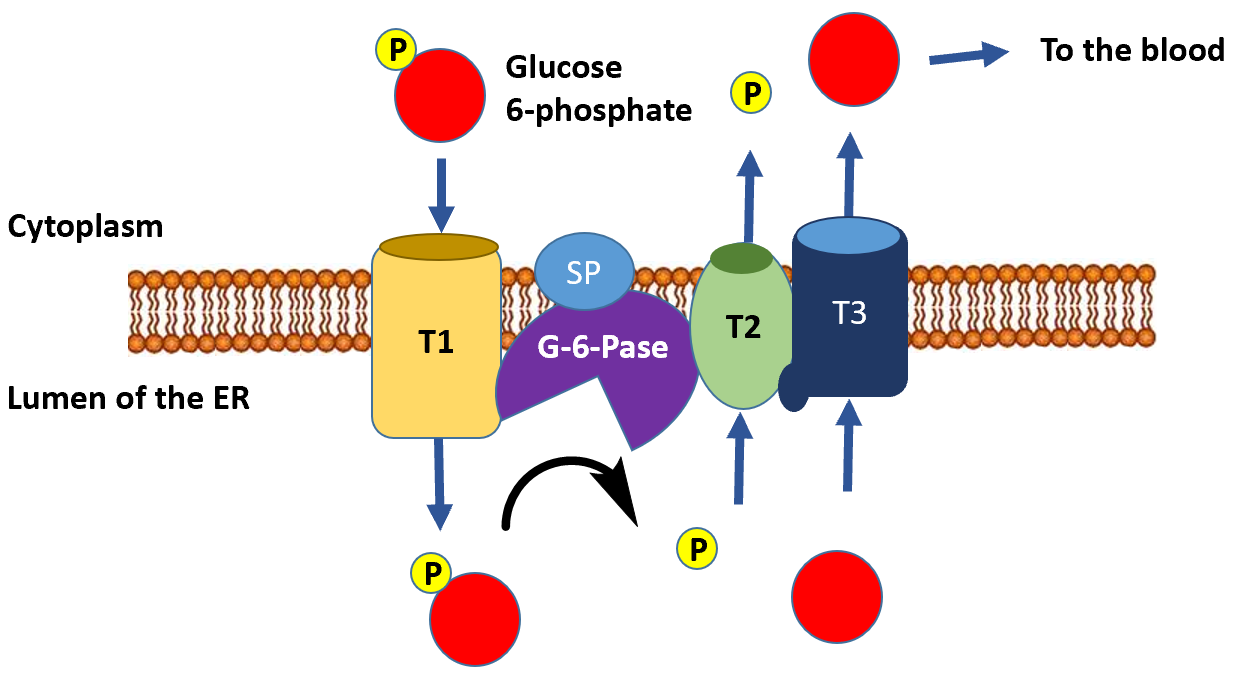
Figure\(\PageIndex{20}\) Dephosphorylation of Glucose 6-Phosphate in the Lumen of the Endoplasmic Reticulum.
The catalytic site of this hydrolase enzyme is comprised of Lys76, Arg83, His119, Arg170, and His176. His 176 acts as a nucleophile that attacks the phosphorous of G6P in an SN2-like reaction to form a His-PO32- intermediate. Hydrolysis follows restoring the enzyme to its original state. Figure \(\PageIndex{21}\) shows an abbreviated mechanism for the reaction.
Figure \(\PageIndex{21}\): Abbreviate mechanism for human glucose-6-phosphatase
Figure \(\PageIndex{21}\) shows an interactive iCn3D model of the AlphaFold predicted structure of human glucose-6-phosphatase (P35575). The enzyme contains nine transmembrane helices that dock the protein in the inner membrane of the ER. The active site faces the ER lumen.
Figure \(\PageIndex{14}\): AlphaFold predicted structure of human glucose-6-phosphatase (P35575)(Copyright; author via source). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i...jmcQAo49eZ8p16
Summary:
It is noteworthy that the process of gluconeogenesis is energy intensive. For example, the phosphatase hydrolysis reactions that liberate glucose are not reverse kinase reactions and they do not produce substrate-level phosphorylation the way reactions occur in the second half of the glycolytic pathway. Overall, there are approximately 6 molar equivalents of ATP required to make 1 mole of glucose. Over 3 times as much energy is consumed during glucose formation than is generated from glycolysis during the breakdown of glucose. Per glucose molecule made, 2 ATP are used in the pyruvate carboxylase step, 2 GTP are used in the phosphoenolpyruvate carboxykinase step, and 2 ATP are used in the kinase reaction to create 1,3-bisphosphoglycerate from 3-phosphoglycerate. There is also the loss of 2 moles of NADH during the reverse reaction to create glyceraldehyde 3-phosphate from 1,3-bisphosphoglycerate, which reduces energy potential through oxidative phosphorylation in the mitochondria. Plus there are energy costs to changing the metabolite pools in the matrix of the mitochondria and for transporting molecules across the mitochondrial membrane that we have not considered here.
References
Principles of Biochemistry/Gluconeogenesis and Glycogenesis. (2019, February 8). Wikibooks, The Free Textbook Project. Retrieved 22:01, August 11, 2022, from https://en.wikibooks.org/w/index.php?title=Principles_of_Biochemistry/Gluconeogenesis_and_Glycogenesis&oldid=3515124.




.png?revision=1&size=bestfit&width=796&height=399)
.png?revision=1&size=bestfit&width=473&height=369)
.png?revision=1&size=bestfit&width=354&height=447)
.png?revision=1&size=bestfit&width=282&height=404)