2.10: Proteins
- Page ID
- 3788
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Proteins are macromolecules. They are constructed from one or more unbranched chains of amino acids; that is, they are polymers. An average eukaryotic protein contains around 500 amino acids but some are much smaller (the smallest are often called peptides) and some much larger (the largest to date is titin a protein found in skeletal and cardiac muscle; one version contains 34,350 amino acids in a single chain!).
Every function in the living cell depends on proteins.
- Motion and locomotion of cells and organisms depends on proteins. [Examples: Muscles, Cilia and Flagella]
- The catalysis of all biochemical reactions is done by enzymes, which contain protein.
- The structure of cells, and the extracellular matrix in which they are embedded, is largely made of protein. [Examples: Collagens] (Plants and many microbes depend more on carbohydrates, e.g., cellulose, for support, but these are synthesized by enzymes.)
- The transport of materials in body fluids depends of proteins.
- The receptors for hormones and other signaling molecules are proteins.
- Proteins are an essential nutrient for heterotrophs.
- The transcription factors that turn genes on and off to guide the differentiation of the cell and its later responsiveness to signals reaching it are proteins.
- and many more — proteins are truly the physical basis of life.
 The protein represented here displays many of the features of proteins. Let's examine some of them as you scroll down the image. The protein consists of two polypeptide chains, a long one on the left of 346 amino acids — it is called the heavy chain — and a short one on the right of 99 amino acids. The heavy chain is shown as consisting of 5 main regions or domains:
The protein represented here displays many of the features of proteins. Let's examine some of them as you scroll down the image. The protein consists of two polypeptide chains, a long one on the left of 346 amino acids — it is called the heavy chain — and a short one on the right of 99 amino acids. The heavy chain is shown as consisting of 5 main regions or domains:
- three extracellular domains, designated here as N (includes the N-terminal), C1, and C2;
- a transmembrane domain where the polypeptide chain passes through the plasma membrane of the cell;
- a cytoplasmic domain (with the C terminal) within the cytoplasm of the cell.

Inteins
Another, very rare, post-translational modification is the later removal of a section of the polypeptide and the splicing together (with a peptide bond) of the remaining N-terminal and C-terminal segments. The portion removed is called an intein (a "protein intron"), and the ligated segments are called exteins ("protein exons"). Genes encoding inteins have been discovered in a variety of organisms, including
- some "true" bacteria such as
- Bacillus subtilis
- several mycobacteria
- several blue-green algae (cyanobacteria)
- some Archaea such as
- Methanococcus jannaschii
- Aeropyrum pernix
- and a few unicellular eukaryotes, e.g., budding yeast (Saccharomyces cerevisiae).
- None has been found in the genomes of multicellular eukaryotes like Drosophila, C. elegans, or the green plant Arabidopsis.
How proteins get their shape
The function of a protein is determined by its shape. The shape of a protein is determined by its primary structure(sequence of amino acids). The sequence of amino acids in a protein is determined by the sequence of nucleotides in the gene (DNA) encoding it. The function of a protein (except when it is serving as food) is absolutely dependent on its three-dimensional structure. A number of agents can disrupt this structure thus denaturing the protein.
- changes in pH (alters electrostatic interactions between charged amino acids)
- changes in salt concentration (does the same)
- changes in temperature (higher temperatures reduce the strength of hydrogen bonds)
- presence of reducing agents (break S-S bonds between cysteines)
None of these agents breaks peptide bonds, so the primary structure of a protein remains intact when it is denatured. When a protein is denatured, it loses its function.

- A denatured enzyme ceases to function.
- A denatured antibody no longer can bind its antigen.
Often when a protein has been gently denatured and then is returned to normal physiological conditions of temperature, pH, salt concentration, etc., it spontaneously regains its function (e.g. enzymatic activity or ability to bind its antigen). This tells us
- The protein has spontaneously resumed its native three-dimensional shape.
- Its ability to do so is intrinsic; no outside agent was needed to get it to refold properly.
However, there are:
- enzymes that add sugars to certain amino acids, and these may be essential for proper folding;
- proteins, called molecular chaperones, that may enable a newly-synthesized protein to acquire its final shape faster and more reliably than it otherwise would.
Chaperones
Although the three-dimensional (tertiary) structure of a protein is determined by its primary structure, it may need assistance in achieving its final shape.
- As a polypeptide is being synthesized, it emerges (N-terminal first) from the ribosome and the folding process begins.
- However, the emerging polypeptide finds itself surrounded by the watery cytosol and many other proteins.
- As hydrophobic amino acids appear, they must find other hydrophobic amino acids to associate with. Ideally, these should be their own, but there is the danger that they could associate with nearby proteins instead — leading to aggregation and a failure to form the proper tertiary structure.
To avoid this problem, the cells of all organisms contain molecular chaperones that stabilize newly-formed polypeptides while they fold into their proper structure. The chaperones use the energy of ATP to do this work.
Chaperonins
Some proteins are so complex that a subset of molecular chaperones — called chaperonins — is needed. Chaperonins are hollow cylinders into which the newly-synthesized protein fits while it folds. The inner wall of the cylinder is lined with hydrophobic amino acids which stabilize the hydrophobic regions of the polypeptide chain while it folds safely away from the
- watery cytosol and
- other proteins outside.
Chaperonins also use ATP as the energy source to drive the folding process.
As mentioned above, high temperatures can denature proteins, and when a cell is exposed to high temperatures, several types of molecular chaperones swing into action. For this reason, these chaperones are also called heat-shock proteins (HSPs). Not only do molecular chaperones assist in the folding of newly-synthesized proteins, but some of them can also unfold aggregated proteins and then refold the protein properly. Protein aggregation is the cause of disorders such as Alzheimer's disease, Huntington's disease, and prion diseases (e.g., "mad-cow" disease). Perhaps some day ways will be found to treat these diseases by increasing the efficiency of disaggregating chaperones.
Despite the importance of chaperones, the rule still holds: the final shape of a protein is determined by only one thing: the precise sequence of amino acids in the protein. And the sequence of amino acids in every protein is dictated by the sequence of nucleotides in the gene encoding that protein. So the function of each of the thousands of proteins in an organism is specified by one or more genes.
Primary Structure

The primary structure of a protein is its linear sequence of amino acids and the location of any disulfide (-S-S-) bridges. Note the amino terminal or "N-terminal" (NH3+) at one end; carboxyl terminal ("C-terminal") (COO-) at the other.
Secondary Structure
Most proteins contain one or more stretches of amino acids that take on a characteristic structure in 3-D space. The most common of these are the alpha helix and the beta conformation.
Alpha Helix

The R groups of the amino acids all extend to the outside.
- The helix makes a complete turn every 3.6 amino acids.
- The helix is right-handed; it twists in a clockwise direction.
- The carbonyl group (-C=O) of each peptide bond extends parallel to the axis of the helix and points directly at the -N-H group of the peptide bond 4 amino acids below it in the helix. A hydrogen bond forms between them [-N-H·····O=C-]
Beta Conformation
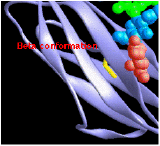 consists of pairs of chains lying side-by-side and
consists of pairs of chains lying side-by-side and- stabilized by hydrogen bonds between the carbonyl oxygen atom on one chain and the -NH group on the adjacent chain.
- The chains are often "anti-parallel"; the N-terminal to C-terminal direction of one being the reverse of the other.
Tertiary Structure
Tertiary structure refers to the three-dimensional structure of the entire polypeptide chain.

The images (courtesy of Dr. D. R. Davies) represent the tertiary structure of the antigen-binding portion of an antibody molecule. Each circle represents an alpha carbon in one of the two polypeptide chains that make up this protein. (The filled circles at the top are amino acids that bind to the antigen.) Most of the secondary structure of this protein consists of beta conformation, which is particularly easy to see on the right side of the image.
Do try to fuse these two images into a stereoscopic (3D) view. I find that it works best when my eyes are about 18" from the screen and I try to relax so that my eyes are directed at a point behind the screen.
Where the entire protein or parts of a protein are exposed to water (e.g., in blood or the cytosol), hydrophilic R groups — including R groups with sugars attached , are found at the surface; hydrophobic R groups are buried in the interior.
Importance of Tertiary structure
The function of a protein (except as food) depends on its tertiary structure. If this is disrupted, the protein is said to be denatured, and it loses its activity. Examples:
- denatured enzymes lose their catalytic power
- denatured antibodies can no longer bind antigen
A mutation in the gene encoding a protein is a frequent cause of altered tertiary structure.
- Curiously, tiny amounts of the mutant version can trigger the alpha-to-beta conversion in the normal protein. Thus the mutant version can be infectious. There have been several cases in Europe of people ill with Creutzfeldt-Jakob disease that may have acquired it from ingesting tiny amounts of the mutant protein in their beef.
- A number of other proteins altered by a point mutation in the gene encoding them, e.g.,
- fibrinogen
- lysozyme
- transthyretin (a serum protein that transports thyroxin and retinol (vitamin A) in the blood)
The many hydrogen bonds that can form between the polypeptide backbones in the beta conformation suggests that this is a stable secondary structure potentially available to many proteins and so a tendency to form insoluble aggregates is as well. Avoidance of amyloid formation may account for the large investment in the cell in
- chaperones
- proteasomes
as well as the crucial importance of particular amino acid side chains in maintaining a globular, and hence soluble, tertiary structure.
Protein Domains
 The tertiary structure of many proteins is built from several domains. Often each domain has a separate function to perform for the protein, such as:
The tertiary structure of many proteins is built from several domains. Often each domain has a separate function to perform for the protein, such as:
- binding a small ligand (e.g., a peptide in the molecule shown here)
- spanning the plasma membrane (transmembrane proteins)
- containing the catalytic site (enzymes)
- DNA-binding (in transcription factors)
- providing a surface to bind specifically to another protein
In some (but not all) cases, each domain in a protein is encoded by a separate exon in the gene encoding that protein. In the histocompatibility molecule shown here ,
- three domains α1, α2, and α3 are each encoded by its own exon.
- Two additional domains a transmembrane domain and a cytoplasmic domainare also encoded by separate exons.
- (β2-microglobulin, "β2m", is NOT a domain of this molecule. It is a separate molecule that binds to the three alpha domains (red line and circle) by noncovalent forces only. The complex of these two proteins is an example of quaternary structure.)
This image (courtesy of P. J. Bjorkman from Nature 329:506, 1987) is a schematic representation of the extracellular portion of HLA-A2, a human class I histocompatibility molecule. It also illustrates two common examples of secondary structure: the stretches of beta conformation are represented by the broad green arrows (pointing N -> C terminal); regions of alpha helix are shown as helical ribbons. The pairs of purple spheres represent the disulfide bridges. A correspondence between exons and domains is more likely to be seen in recently-evolved proteins. Presumably, "exon shuffling" during evolution has enabled organisms to manufacture new proteins, with new functions, by adding exons from other parts of the genome to encode new domains (rather like Lego® pieces).
Quaternary Structure
Complexes of 2 or more polypeptide chains held together by noncovalent forces (usually) but in precise ratios and with a precise 3-D configuration. The noncovalent association of a molecule of beta-2 microglobulin with the heavy chain of each class I histocompatibility molecule is an example.
Protein Kinesis
All proteins are synthesized by ribosomes using the information encoded in molecules of messenger RNA (mRNA). The various destinations for proteins occur in two major sets:
- one set for those proteins synthesized by ribosomes that remain suspended in the cytosol, and
- a second set for proteins synthesized by ribosomes that are attached to the membranes of the endoplasmic reticulum (ER) forming "rough endoplasmic reticulum" (RER).
Some of the important destinations for proteins are:
- the cytosol
- the nucleus
- mitochondria
- chloroplasts
- peroxisomes


