2.3: Genetic Code and Translation
- Page ID
- 32272
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Learning Objectives
- Identify the key steps of translation and the role of tRNAs, aminoacyl tRNA synthetases, and ribosomal RNAs.
- Use the codon table to determine the sequence of amino acids that will be produced from a DNA or mRNA sequence.
- Use the codon table to determine which amino acid will be carried by a tRNA, given the anticodon sequence.
- Identify untranslated regions and include these regions in gene diagrams.
- Explain how some antibiotics affect ribosomes to kill bacteria.
Overview of translation
Genes contain the instructions a cell needs to make proteins. Making proteins from DNA requires a two-step process:
- Transcription: the process of copying the gene’s DNA into RNA
- Translation: the process of using RNA to synthesize a sequence of amino acids to form a polypeptide (or protein)
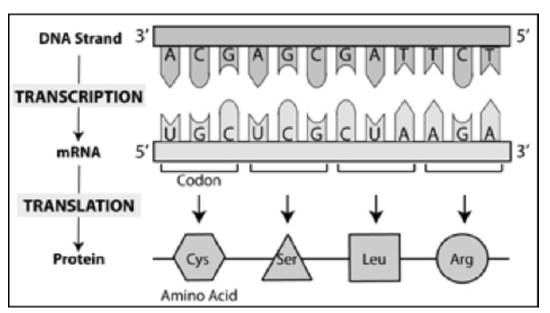
Figure \(\PageIndex{1}\). Summary of transcription and translation showing the relationship of DNA and RNA nucleotides with amino acids (CC BY-NC-SA)
Important Players in Translation
messenger RNA(mRNA): RNA copy of DNA that contains the instructions to make a protein
transfer RNA (tRNA): RNA molecule responsible for delivering amino acids to the ribosome
amino acids: The basic building block of a protein. There are 20 different amino acids, each has an amino group (NH2) and a carboxyl group (COOH), but differs in its functional group (also known as side chain or R group). An infographic showing all the amino acids can be found at this link (https://www.compoundchem.com/wp-content/uploads/2014/09/20-Common-Amino-Acids-Physiological-Structures.pdf).
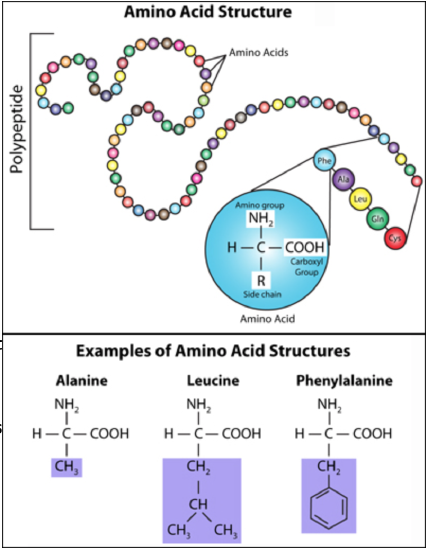
Figure \(\PageIndex{2}\). (CC BY-NC-SA)
protein: A chain of amino acids, also known as a polypeptide; some proteins require multiple polypeptides to function.
ribosome: The organelle on which mRNA is translated into protein. Consists of a large (60S) and small subunit (40S) which are made up of ribosomal RNA (rRNA) and proteins.
The Genetic Code: Translating RNA into Protein
How does the nucleotide sequence of RNA specify the specific order of amino acids in a protein? The answer lies in what is known as the genetic code.
- If each RNA base codes for just 1 amino acid, RNA could code for only 4 amino acids (not enough to include all 20 amino acids).
- If two RNA bases code for 1 amino acid, RNA could code for 16 amino acids (still not enough to include all 20 amino acids).
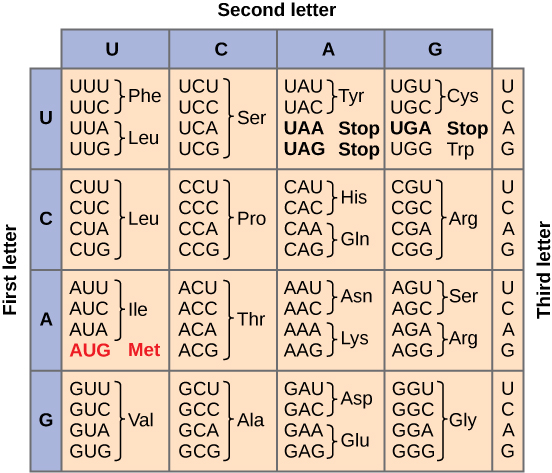
- If three RNA bases code for 1 amino acid, RNA could code for 64 amino acids (more than enough to include all 20 amino acids). Thus, the genetic code is a triplet code in which three nucleotides in RNA specify one amino acid in protein.
In the 1960s, the mRNA code for each amino acid was determined experimentally; today, tables are used to find the amino acid encoded by each codon. Remember that each codon must be read 5' to 3'.
The role of transfer RNA
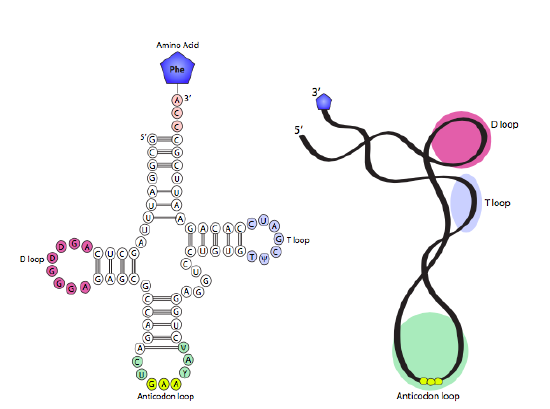
Consider nucleic acids and amino acids as different languages. DNA and RNA speak the language of nucleic acids (A, U/T, G and C); whereas proteins speak the language of amino acids. The tRNA (transfer RNA) is the "interpreter" in the cell because it is the molecule that speaks both languages!
Sets of three nucleotides that code for a specific amino acid are known as codons. These codons are recognized by, and base pair with, the tRNA molecule with the complementary sequence called anticodon. tRNA molecules act as translators because they are able to read the nucleic acid words (as mRNA codons) and deliver the corresponding words (as amino acids). There is at least one tRNA for each of the 20 amino acids (some amino acids bind to 2 or 3 different tRNAs, so cells may contain as many as 32 different tRNAs). The secondary structure of the tRNA relies on complementary sequences within a single tRNA to fold into a function structure with the anticodon loop at the fold opposite the amino acid on the 3' end. Enzymes called aminoacyl-tRNA synthetases catalyze the attachment of the correct amino acid to each tRNA, this process is known as tRNA charging.
Exercise \(\PageIndex{1}\)
A protein is being translated from an mRNA with the sequence 5' augcaaucauaugcuucugcuauguuaagc.. 3'. Starting with aug, what are the first five amino acids of the peptide?
- Answer
-
Met - Gln - Ser - Tyr - Ala
Exercise \(\PageIndex{2}\)
A DNA sequence is 5' ...ATGCCGTATAGG... 3'.
- If this strand is the coding strand, determine the RNA and possible amino acid sequence. (Assume you begin translation at the 5' nucleotide.)
- If this strand is the non-coding (template) strand, determine the RNA and possible amino acid sequence. (Assume you begin translation at the 5' nucleotide.)
- Answer
-
- The RNA has the same sequence as the coding strand, except for U instead of T. The mRNA will be 5' ...AUGCCGUAUAGG... 3' and the protein sequence will be Met - Pro - Tyr - Arg.
- The RNA has a complementary sequence to the template strand. (Remember that transcription would occur from right to left off this template to make RNA from 5' to 3'.) The mRNA will be 5' ...CCUAUACGGCAU... 3' and the protein sequence will be Pro - Ile - Arg - His.
Exercise \(\PageIndex{3}\)
If a tRNA has the anticodon sequence 5' UAC 3', what amino acid will it be carrying?
- Answer
-
Valine (val)
To see this how this problem is solved, watch this video:
The Steps of Translation
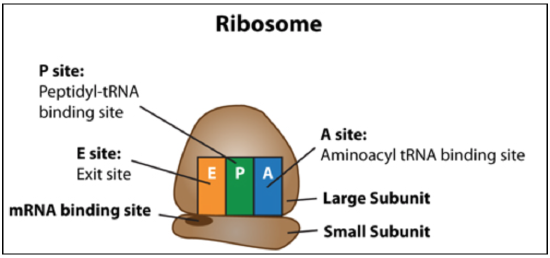
Figure \(\PageIndex{5}\): Summary of ribosome structure and positions of E, P, and A sites. (CC BY-NC-SA)
Translation Initiation: The small subunit binds to a site upstream (on the 5' side) of the start of the mRNA. It proceeds to scan the mRNA in the 5'-to-3' direction until it encounters the START codon (AUG). Nearby sequences help position the small subunit. In prokaryotes, this sequence, called the Shine-Dalgarno sequence, is upstream of the AUG. In eukaryotes, a consensus sequence (Kozak sequence) surrounds the AUG.
When the small subunit and mRNA are correctly positioned with a tRNA, the large subunit attaches and the initiator tRNA, which carries methionine (Met), binds to the P site on the ribosome.
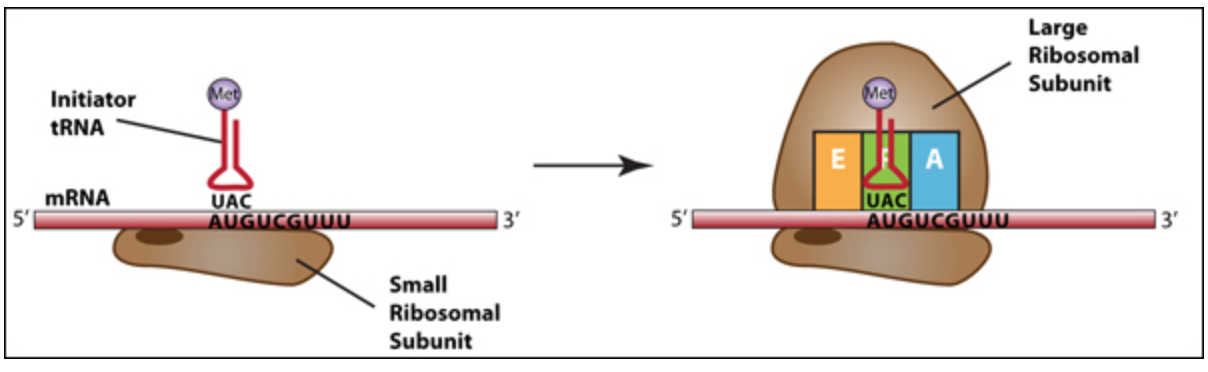
Figure \(\PageIndex{6}\). Translation Initiation: The assembly of the small ribosomal subunit, mRNA and initiator tRNA is followed by the addition of the large ribosomal subunit. The initiator tRNA has sequence UAC that is complementary to AUG in the mRNA. (CC BY-NC-SA)
Elongation: A tRNA bound to its amino acid (known as an aminoacyl-tRNA) that is able to base pair with the next codon on the mRNA arrives at the A site. The ribosome catalyzes the formation of a peptide bond between the preceding amino acid (Met at the start of translation) in the P site and the amino acid held by the tRNA in the A site. The initiator tRNA moves to the E site and the ribosome translocates (moves down) one codon downstream. This shifts the more most recent tRNA from the A site to the P site, opening up the A site for the arrival of a new aminoacyl-tRNA. Polypeptide synthesis repeats, the tRNA residing in the E site is released from the complex, the tRNAs in the P site and A site shift over and the next amino acid is added to the growing polypeptide chain. This cycle repeats until a stop codon is reached.
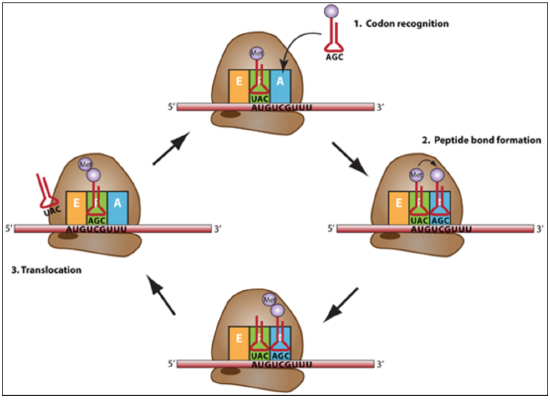
Figure \(\PageIndex{7}\): The elongation cycle of translation begins with codon recognition in the A site by a tRNA with a complementary anticodon, then peptide bond formation and transfer of the amino acid to the A site tRNA, then translocation of the ribosome toward the 3 end of the mRNA for the next codon to be in the A site. (CC BY-NC-SA)
Remembering what happens in the E, P, and A sites
- P = peptidyl because the P site holds the tRNA with the growing peptide
- A = aminoacyl because the A site holds the incoming amino acid that will accept the transfer of the peptide from the P site
- E = exit site because the tRNA from the P site will exit the ribosome through this site after peptide bond formation and translocation
Termination: Translation ends when the ribosome reaches a STOP codon (UAA, UAG or UGA). There are no tRNA molecules with anticodons complementary to stop codons, instead protein release factors (RF) recognize these codons when they arrive at the A site. Binding of a release factor causes the polypeptide (protein) to be released from the ribosome. The ribosome subunits dissociate (split) from each other and can be reassembled later for another round of protein synthesis.
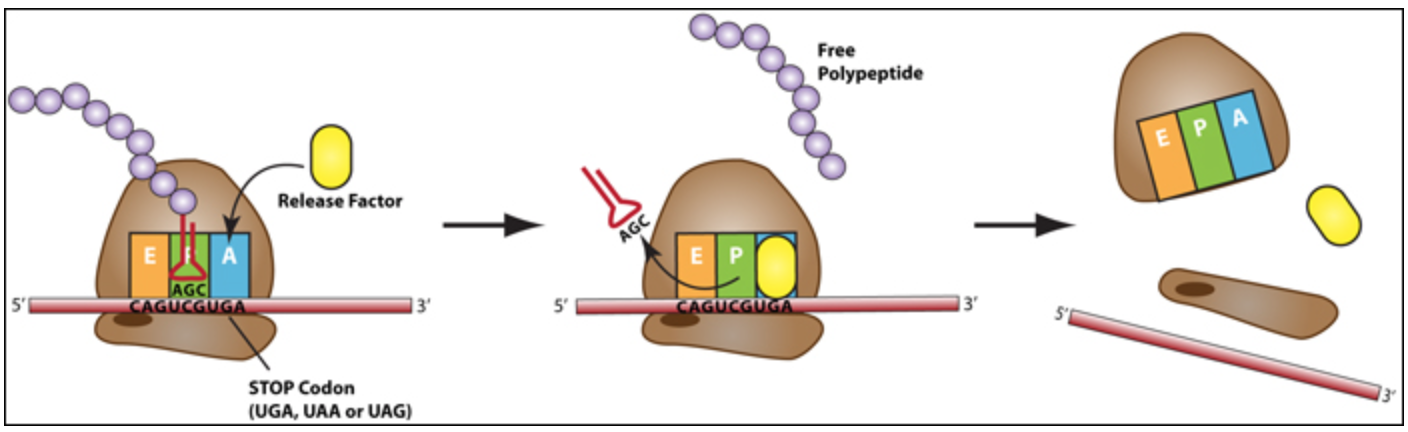
Figure \(\PageIndex{8}\): During termination release factor proteins enter the A site and the small and large ribosomal subunits dissociate from each other and the mRNA. The newly made polypeptide is released. (CC BY-NC-SA)
Translation tutorial modified from:

Translation Tutorial by Dr. Katherine Harris is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License.
Funded by the U.S. Department of Education, College Cost Reduction and Access (CCRAA) grant award # P031C080096.
Untranslated regions (UTRs)
In both prokaryotes and eukaryotes, the start codon for translation (AUG) is not at the +1 site where transcription begins. Similarly, the stop codon for translation is not at the 3' end of the mRNA. This genetic architecture leads to two regions of the mRNA that are called the 5' and 3' untranslated regions (UTRs). To understand what a UTR is, review these key steps of translation initiation and termination.
- For initiation, the small ribosomal subunit moves along the mRNA from 5' to 3' until it is positioned at the start codon. Then the large subunit load onto the complex with the initiator tRNA carrying methionine. Then the ribosome translates the RNA in the 5' to 3' direction, therefore all the nucleotides prior to the start codon were skipped, or untranslated.
- For termination, when the ribosome encounters a stop codon, there is not a tRNA with a complementary codon. Instead release factor proteins bind in that site and disrupt the complex, causing the mRNA and ribosome to disassociate. Therefore, any nucleotides after the stop codon cannot be translated, because the ribosome no longer moves along this part of the mRNA.
Example \(\PageIndex{1}\)
Consider a relatively small gene, the SRY gene on the human Y chromosome. The SRY mRNA is 828 nucleotides. If each amino acid is coded by 3 nucleotides (nt) and the entire mRNA encoded amino acids, the translated protein would be 276 amino acids.
However, the SRY protein is 204 amino acids. How many nucleotides are in the untranslated regions?
Solution
If 1 amino acid is encoded by 3 nucleotides, then 204 amino acids x 3 nucleotides = 612 nucleotides to encode a 204 amino acid protein. Adding 3 nucleotides for the stop codon adds 3 more nucleotides (612 + 3) = 615 nucleotides of coding sequence.
The SRY mRNA is 828 nucleotides, so the remaining (828 - 615) 213 nucleotides must be in untranslated regions. The nucleotides upstream (5' of) the AUG start codon are the 5' UTR and the nucleotides downstream of the stop codon (in this gene, UAG) are the 3' UTR.
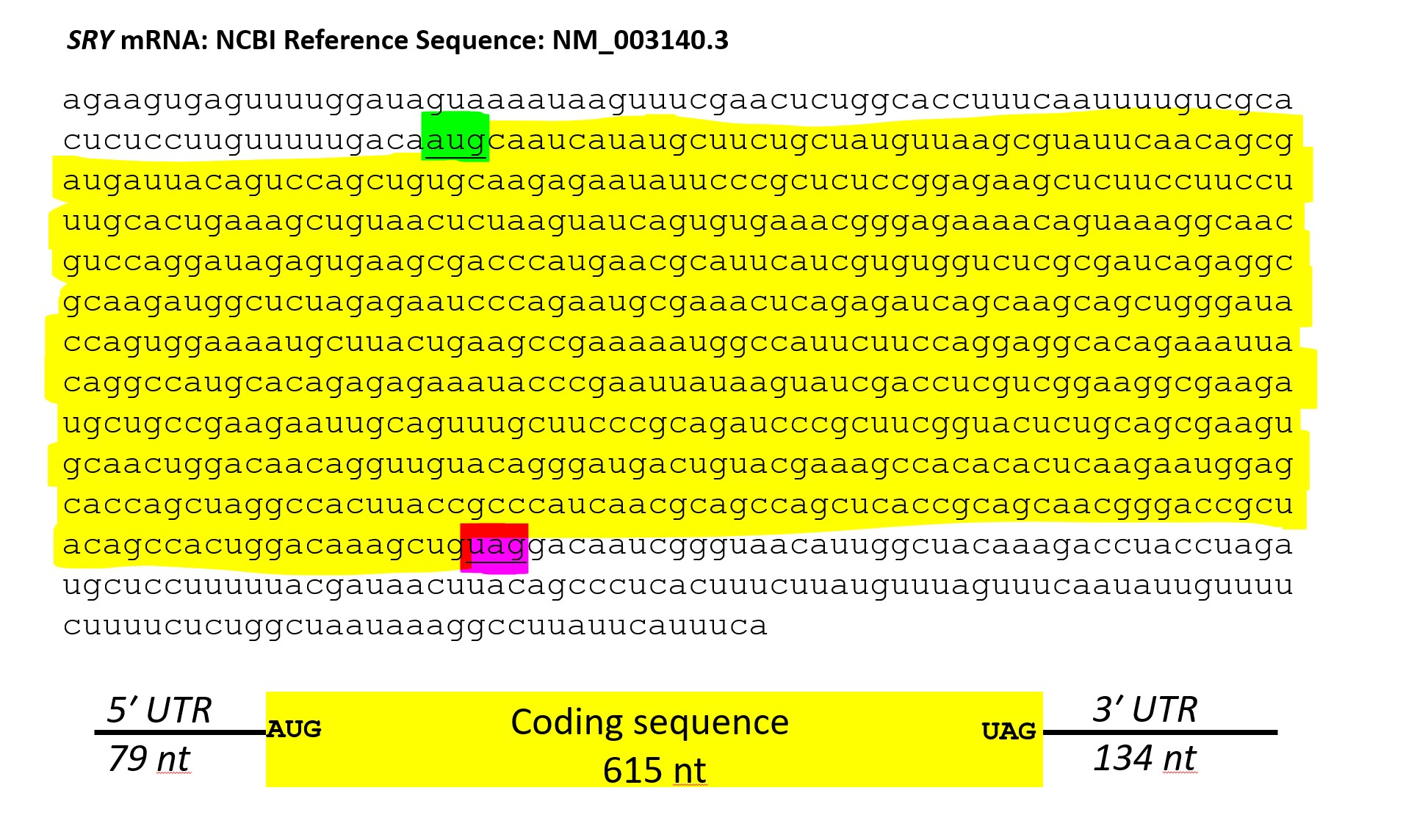
Why have untranslated regions?
Untranslated regions often function in RNA stability, translation initiation, or other aspects of post-transcriptional regulation; however the complete function of most untranslated regions is not entirely known.
The importance of ribosomal RNA
The ribosome itself is made of both ribosomal proteins and rRNA (ribosomal RNA). In a Perspective from 2000 titled "The Ribosome is a Ribozyme," (https://science.sciencemag.org/content/289/5481/878) Thomas Cech describes how the crystal structure of the ribosome advanced our understanding of the roles of the rRNA and protein components of this critical complex. The crystal structure of the large ribosomal subunit from the archean Haloarcula marismortui at high resolution was published in 2000 (https://science.sciencemag.org/content/289/5481/905 Ban et al.). Venkatraman Ramakrishnan, Thomas A. Steitz and Ada E. Yonath later shared the 2009 Nobel Prize in Chemistry for their contributions to our understanding of this structure. This structure revealed that the active site, where the peptide bond forms between the amino acids in the P and A sites, is largely not proteins, which most cellular catalysts are, but instead ribosomal RNA. A ribozyme is the term for an RNA that can catalyze an enzymatic reaction. Therefore, the ribosome is a ribozyme.
Structure of the 50S ribosomal subunit assembly (intermediate state 1) from E. coli K-12 (https://www.rcsb.org/structure/6GC7) with ribosomal RNA in orange and proteins in other colors.
Antibiotics and the ribosome
In general, protein synthesis inhibitors work at different stages of prokaryotic mRNA translation into proteins like initiation, elongation (including aminoacyl tRNA entry, proofreading, peptidyl transfer, and ribosomal translocation), and termination. The following is a list of common antibacterial drugs and the stages which they target.
- Linezolid acts at the initiation stage, probably by preventing the formation of the initiation complex, although the mechanism is not fully understood.
- Tetracyclines and Tigecycline (a glycylcycline related to tetracyclines) block the A site on the ribosome, preventing the binding of aminoacyl tRNAs.
- Aminoglycosides, among other potential mechanisms of action, interfere with the proofreading process, causing an increased rate of error in synthesis with premature termination.
- Chloramphenicol blocks the peptidyl transfer step of elongation on the 50S ribosomal subunit in both bacteria and mitochondria.
- Macrolides, clindamycin, and aminoglycosides have evidence of inhibition of ribosomal translocation.
- Streptogramins also cause premature release of the peptide chain.
By targeting different stages of the mRNA translation, antimicrobial drugs can be changed if resistance develops to one or many of the drugs.
Contributor:
- General Microbiology Boundless from: https://bio.libretexts.org/Courses/Northwest_University/MKBN211%3A_Introductory_Microbiology_(Bezuidenhout)/07%3A_Antimicrobial_Drugs/7.02%3A_Functions_of_Antimicrobial_Drugs/7.2.04%3A_Inhibiting_Protein_Synthesis
Thinking about antibiotics and specificity
Antibiotics are used to treat bacterial infections. Consider these questions:
- Why are antibiotics not prescribed for viral infections?
- If humans have ribosomes as well, why would the antibiotics not inhibit the human ribosomes and prevent our own protein production?
- What does antibiotic resistance mean and how does it arise?
- Does the same antibiotic treat all types of bacterial infections? Why or why not?
- How does studying the structure of the ribosome help us to understand the effectiveness of antibiotics?


