10.5: tRNA are Rather Odd Ducks
- Page ID
- 16153
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)In prokaryotes, tRNA can be found either as single genes or as parts of operons that can also contain combinations of mRNAs or rRNAs. In any case, whether from a single gene, or after the initial cleavage to separate the tRNA transcript from the rest of the transcript, the resulting pre-tRNA has an N-terminal leader (41 nt in E. coli) that is excised by RNase P. That cleavage is universal for any prokaryotic tRNA. After that, there are variations in the minor excisions carried out by a variety of nucleases that produce the tRNA in its final length though not its mature sequence, as we will see in a few paragraphs.
Eukaryotic pre-tRNA (transcribed by RNA polymerase III) similarly has an N-terminal leader removed by RNase P. Unlike the prokaryote though, the length can vary between different tRNAs of the same species. Some eukaryotic pre-tRNA transcripts also contain introns, especially in the anticodon loop, that must be spliced out for the tRNA to function normally. These introns are different from the self-splicing or spliceosome- spliced transcripts discussed in the transcription chapter. Here, the splicing function is carried out not by ribozymes, but by conventional (protein) enzymes. Interestingly, RNaseP also removes a 3’ sequence from the pre-tRNA, but then another 3’ sequence is added back on. This new 3’ end is always CCA, and is added by three successive rounds with tRNA nucleotidyl transferase.
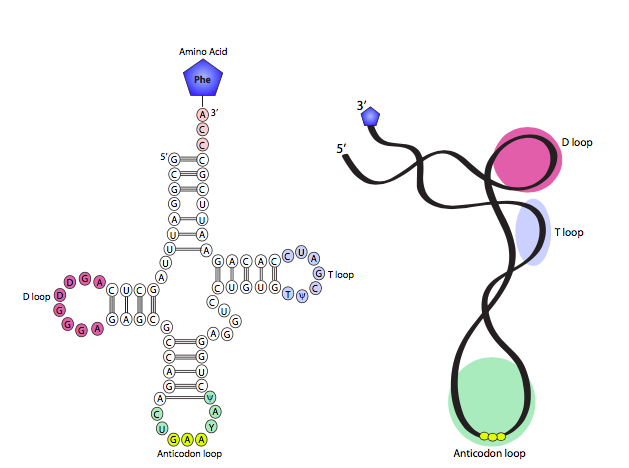
When RNA was introduced, it was noted that although extremely similar to DNA on many counts, it is normally single stranded, and that property, combined with the opportunity for complementary base pairing within a strand, allows it to do something far different than double-stranded DNA: it can form highly complex secondary structures. One of the simplest and clearest examples of this is tRNA, which depends on its conformation to accomplish its cellular function. The prototypical cloverleaf-form tRNA diagram is shown in Figure \(\PageIndex{2}\) on the left, with a 3D model derived from x-ray crystallographic data on the right. As you can see, the fully-splayed-out shape has four stem-loop “arms” with the amino acid attached to the acceptor arm, which is on the opposite side of the tRNA from the anticodon arm, which is where the tRNA must match with the mRNA codon during translation. Roughly perpendicular to the acceptor-anticodon axis are the D arm and the TyC arm. In some tRNAs, there are actually five total arms with a very short loop between the TyC and anticodon arms. The arm-like stem and loop structures are formed by two areas of strong complementarity (the stems, base-paired together) interrupted by a short non-complementary sequence (the loop). In general terms, the arms are used to properly position the tRNA within the ribosome as well as recognizing the mRNA codon and bringing in the correct amino acid.
When it comes time for the tRNA to match its anticodon with the codon on the mRNA, the code is not followed “to the letter” if you will pardon the pun. There is a phenomenon called “wobble” in which a codon-anticodon match is allowed and stabilized for translation even if the nucleotide in the third position is not complementary. Wobble can occur because the conformation of the tRNA allows a little flexibility to that position of the anticodon, permitting H-bonds to form where they normally would not. This is not a universal phenomenon though: it only applies to situations where a U or a G is in the first position of the anticodon (matching the third position of codon). Following the convention of nucleic acid sequences, the sequence is always written 5’ to 3’, even though in the case of codon-anticodon matching, the strands of mRNA and tRNA are antiparallel:
In addition to being allowed a bit of wobble in complementary base-pairing, tRNA molecules have another peculiarity. After being initially incorporated into a tRNA through conventional transcription, there is extensive modification of some of the bases of the tRNA. This affects both purines and pyrimidines, and can range from simple additions such as methylation or extensive restructuring of the sugar skeleton itself, as in the conversion of guanosine to wyosine (W). Over 50 different modifications have been catalogued to date. These modifications can be nearly universal, such as the dihydrouridine (D) found in the tRNA D loop, or more specific, such as the G to W conversion found primarily in tRNAPhe of certain species (examples have been identified in both prokaryotic and eukaryotic species). As many as 10% of the bases in a tRNA may be modified. Naturally, alterations to the sugar base of the nucleotides can also alter the base-pairing characteristics. For example, one common modified base, inosine, can complement U, C, or A. This aberrant complementary base pairing can be equal among the suitor bases, or it may be biased, as in the case of 5-methoxyuridine, which can recognize A, G, or U, but the recognition of U is poor.
Charging the tRNA
The knowledge of the genetic code begs the question: how is the correct amino acid attached to any given tRNA? A class of enzymes called the aminoacyl tRNA synthetases are responsible for recognizing both a specific tRNA and a specific amino acid, binding an ATP for energy and then joining them together (sometimes called "charging the tRNA") with hydrolysis of the ATP. Specificity is a difficult task for the synthetase since amino acids are built from the same backbone and are so similar in mass. Distinguishing between tRNA molecules is easier, since they are larger and their secondary structures also allow for greater variation and therefore greater ease of discrimination. There is also a built-in pre-attachment proofreading mechanism in that tRNA molecules that fit the synthetase well (i.e. the correct ones) maintain contact longer and allow the reaction to proceed whereas ill-fitting and incorrect tRNA molecules are likely to disassociate from the synthetase before it tries to attach the amino acid.
Charging an aminoacyl tRNA synthetase with its amino acid requires energy. The synthetase first binds a molecule of ATP and the appropriate amino acid, which react resulting in the formation of aminoacyl-adenylate and pyrophosphate. The PPi is released and the synthetase now binds to the proper tRNA. Finally, the amino acid is transferred to the tRNA. Depending on the class of synthetase, the amino acid may attach to the 2’-OH of the terminal A (class I) or to the 3’-OH of the terminal A (class II) of the tRNA. Phe-tRNA synthetase is the exception: it is structurally a class II enzyme but transfers the Phe onto the 2’-OH. Note that amino acids transferred onto the 2’-OH are soon moved to the 3’-OH anyway due to a transesterification reaction.


