2.2: RNA processing
- Page ID
- 32271
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Learning Objectives
- List three mRNA-processing events that occur after transcription in eukaryotes and identify the purpose of each process.
- Explain how alterative splicing produces multiple distinct mRNAs from a single gene.
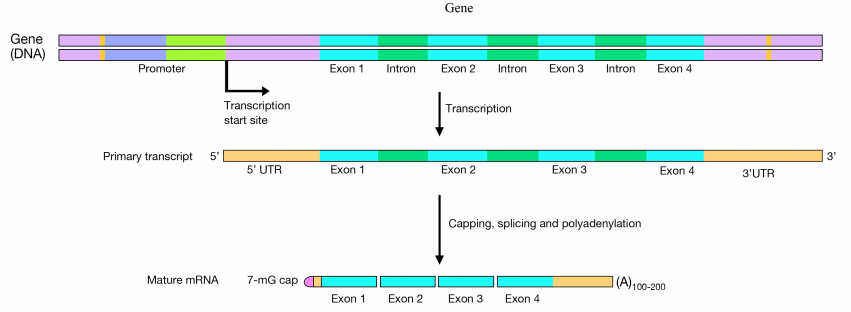
mRNA processing in eukaryotes
Transcription is the mechanism by which the information in genes (DNA) is used to produce RNA. The newly-made RNA, also known as the primary transcript (the product of transcription is known as a transcript) or pre-mRNA, is further processed before it is functional. Transcripts that encode proteins will proceed to the ribosome for translation. In bacterial cells, the mRNA can be translated directly as it comes off the DNA template. In eukaryotic cells, RNA synthesis, which occurs in the nucleus, is separated from the protein synthesis machinery, which is in the cytoplasm. In addition, eukaryotic genes have introns, which are non-coding regions that interrupt the gene’s coding sequence. The primary RNA copied from genes containing introns will also therefore have regions that interrupt the coding sequence of the gene. These regions must be removed before the mRNA is sent out of the nucleus to be used to direct protein synthesis. The process of removing the introns and rejoining the coding sections or exons, of the mRNA, is called splicing. Proteins that interact with the cap, mRNA, and poly(A) tail allow the mature mRNA to be exported through nuclear pores into the cytoplasm for translation.
What are the processing steps for messenger RNAs?
In eukaryotic cells, pre-mRNAs undergo three main processing steps:
- Capping at the 5' end
- Addition of a poly(A) tail at the 3' end
- Splicing to remove introns
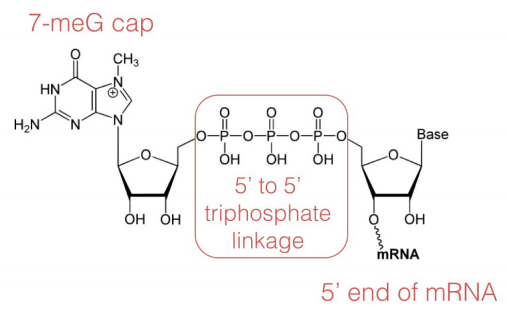
Capping
In the capping step of mRNA processing, a methylated-guanosine (7-methyl-G) is linked to the phosphates at the 5' end of the mRNA. The cap protects the 5' end of the mRNA from degradation by nucleases and also helps to position the mRNA correctly on the ribosomes during protein synthesis.
Poly(A) tail addition
The 3' end of a eukaryotic mRNA is first trimmed, then an enzyme called Poly(A) Polymerase adds a "tail" of about 200 ‘A’ nucleotides to the 3' end. There is evidence that the poly(A) tail plays a role in efficient translation of the mRNA, as well as in the stability of the mRNA. The cap and the poly(A) tail on an mRNA indicate that the mRNA is complete (i.e., not defective).
Splicing
In many cases, splicing now appears to occur as the transcript is being synthesized. Introns are removed from the pre-mRNA by the activity of a complex called the spliceosome. The spliceosome is made up of proteins and small nuclear RNAs (snRNAs) that associate to form protein-RNA enzymes called small nuclear ribonucleoproteins or snRNPs (pronounced SNURPS). The splicing machinery must be able to recognize sequences that are specific to splice junctions (i.e., the end of each exon and the start of the next) in order to correctly cut out the introns and join the exons together to make the mature, spliced mRNA.
Introns vs Exons: Which one is "in" ?
- The word intron comes from the word intervening; these are the intervening sequences that break up the coding sequence. You can remember that introns are "in the way" of the coding sequence.
- The word exon comes from the word expressed; these sequences are present in the mature mRNA.
What signals indicate where an intron starts and ends? The base sequence at the start (5' or left end, also called the splice donor or donor site) of an intron is GU while the sequence at the 3' or right end (also called the splice acceptor or acceptor site) is AG. There is also a third important sequence within the intron, called a branch point, that is important for splicing. These sequences interact with the snRNAs
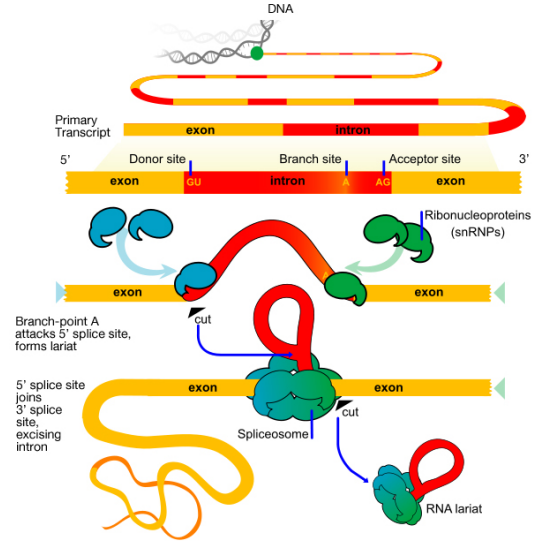
There are two main steps in splicing:
- In the first step, the pre-mRNA is cut at the 5' splice site (the junction of the 5' exon and the intron). The 5' end of the intron then is joined to the branch point within the intron. This generates the lariat-shaped molecule characteristic of the splicing process
- In the second step, the 3' splice site is cut, and the two exons are joined together, and the intron is released.
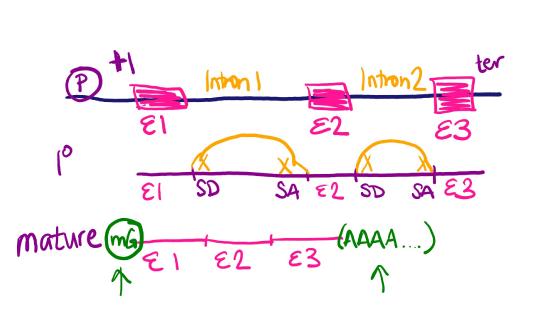
Exercise \(\PageIndex{1}\)
Draw a diagram of a section of DNA that has a gene with three exons and two introns. Draw the corresponding primary transcript and mature mRNA. Label the promoter, transcription start site, exons, introns, splice donors (SD) and splice acceptors (SA), transcription termination, 5' cap, and polyA tail.
- Answer
-
Your gene diagram should look something like this:
For a walk-through of creating the diagram, you can view this video:
Exercise \(\PageIndex{2}\)
Where does eukaryotic mRNA processing occur?
A. All three steps occur in the nucleus.
B. All three steps occur in the cytoplasm.
C. The cap and tail are added in the nucleus, but splicing occurs in the cytoplasm.
D. Splicing occurs in the nucleus, but the cap and tail are added in the cytoplasm.
- Answer
-
A.
Alternative Splicing
Many pre-mRNAs have a large number of exons that can be spliced together in different combinations to generate different mature mRNAs. This is called alternative splicing, and allows the production of many different proteins using relatively few genes, because a single RNA can, by combining different exons during splicing, create many different protein coding messages. Because of alternative splicing, each gene in our DNA gives rise, on average, to three different proteins.
Alternative splicing may also be used to change or downregulate protein production from a gene, by retaining and intron that includes a premature stop codon, which will end protein synthesis before the final protein can be completed. This role for alternative splicing may be useful for organisms that need to respond to a changing environment, such as temperature or light for plants. Profiling of mRNAs after cold treatment in tea plants revealed that 28.8% of alternatively spliced genes retained an intron (Li et al, 2020).
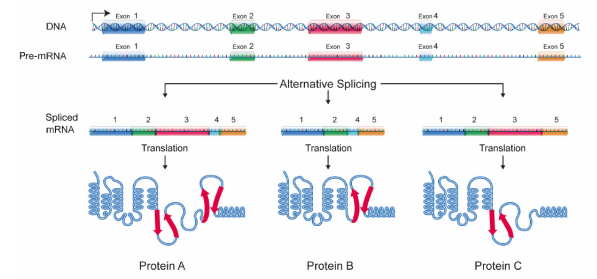
Figure \(\PageIndex{4}\): Splicing and protein diversity
tRNAs and rRNAs are Processed After Transcription
In addition to mRNA processing, non-coding eukaryotic RNAs are processed (trimmed, chemically modified) from large precursor RNAs to mature, functional RNAs. These precursor RNAs (pre-RNAs, or primary transcripts) contain in their sequences the information necessary for their function in the cell. Both prokaryotes and eukaryotes process their ribosomal and transfer RNAs.
- Pre-rRNA (ribosomal RNA) is cleaved and/or trimmed (not spliced because the pieces become separate rRNAs) to make multiple, shorter mature rRNAs.
- Pre-tRNAs (transfer RNA) are trimmed, some bases within the transcript are chemically modified and 3' bases (not encoded by the tRNA gene) are enzymatically added to the 3'-end.
References
Li, Y., Mi, X., Zhao, S. et al. Comprehensive profiling of alternative splicing landscape during cold acclimation in tea plant. BMC Genomics 21, 65 (2020). https://doi.org/10.1186/s12864-020-6491-6
Zahler AM. Alternative splicing in C. elegans. WormBook. 2005 Sep 26:1-13. doi: 10.1895/wormbook.1.31.1. PMID: 18050427; PMCID: PMC4781185.
Contributors and Attributions
Dr. Kevin Ahern and Dr. Indira Rajagopal (Oregon State University)
- Gerald Bergtrom

