B. Translation - First Steps
1. Making Aminoacyl-tRNAs
Translation is perhaps the most energy-intensive job a cell must do, beginning with the attachment of amino acids to their tRNAs. The basic amino-acylation reaction is the same for all amino acids. A specific aminoacyl-tRNA synthase attaches each tRNA to (charges) an appropriate amino acid. Charging tRNAs requires ATP and proceeds in three steps (shown below).
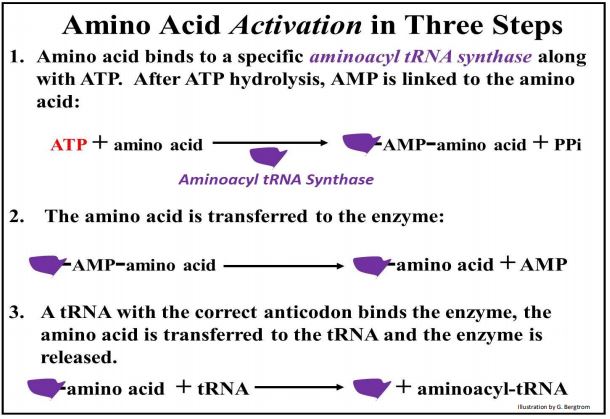
In the first step, ATP and an appropriate amino acid bind to the aminoacyl-tRNA synthase. ATP is hydrolyzed releasing a pyrophosphate (PPi) and leaving an enzyme-AMP-amino acid complex. Next, the amino acid is transferred to the enzyme, releasing the AMP. Finally, the tRNA binds to the enzyme, the amino acid is transferred to the tRNA and the intact enzyme is regenerated and released. The charged tRNA is ready to use in translation.
Several studies had already established that polypeptides are synthesized from their amino (N-) terminal end to their carboxyl (C-) terminal end. When it became possible to determine the amino acid sequences of polypeptides, it turned out that around 40% of E. coli proteins had an N-terminal methionine, suggesting that all proteins began with a methionine. It also turned out that, even though there is only one codon for methionine, two different tRNAs for methionine could be isolated. One of the tRNAs was bound to a methionine modified by formylation, called formylmethionine-tRNAfmet (or fmet-tRNAf for short). The other was methionine-tRNAmet (met-tRNAmet for short), charged with an unmodified methionine.
Methionine and formylated methionine are shown below.
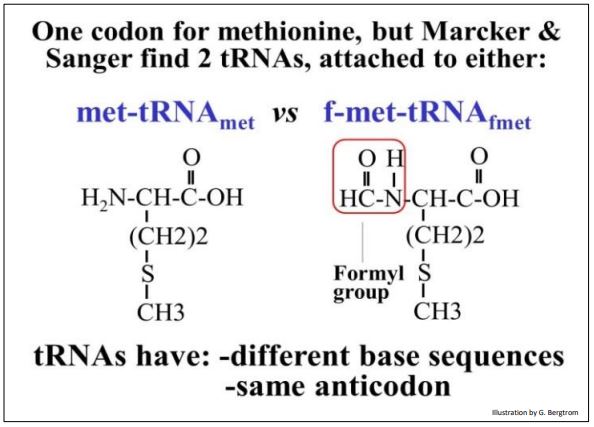
tRNAmet and tRNAf each have an anticodon to AUG, the only codon for methionine, but have different base sequences encoded by different tRNA genes. tRNAmet is used to insert methionine in the middle of a polypeptide. tRNAf is the initiator tRNA, and is only used to start new polypeptides with formylmethionine. In prokaryotes, methionine on met-tRNAf is formylated at its amino group to make the fmet-tRNAf. The formylating enzyme that does this does not recognize methionine on met-tRNAmet.
In E. coli, a formylase enzyme removes the formyl group from all N-terminal formyl methionines at some point after translation has begun. As we noted, the methionine itself (and sometimes more N-terminal amino acids) are also removed from about 60% of E. coli polypeptides. Eukaryotes have inherited both the initiator tRNAf and the tRNAmet, using only met-tRNAf during initiation. However, methionine on the eukaryotic initiator met-tRNAf is never formylated in the first place. What’s more, methionine is absent from virtually all mature eukaryotic polypeptides.
Early in evolution, the need for an initiator tRNA must have ensured a correct starting point for translation on an mRNA and therefore growth of a polypeptide from one end to the other, that is, from its N- to its C-terminus. At one time, formylation of the N-terminal methionine may have served to block accidental addition of amino acids or other modifications at the N-terminus of a polypeptide. Today, formylation seems to be a kind of molecular appendix in bacteria. Since then, evolution (in eukaryotes at least) has selected other features to replace actual formylation as the protector of the N-terminus of polypeptides.
2. Initiation
Now that we have charged the tRNAs, we can look more closely at the three steps of translation. Understanding translation initiation began with a molecular dissection of the components of E. coli cells required for cell-free (in vitro) protein synthesis, including cell fractionation, protein purification and reconstitution experiments. Initiation starts with when the Shine-Delgarno sequence forms Hbonds with a complementary sequence in the 16S rRNA bound to 30S ribosomal subunit. The Shine-Delgarno sequence is a short nucleotide sequence in the 5’ untranslated region (5’-UTR) of the messenger RNA, just upstream of the initiator AUG codon. This requires the participation of initiation factors IF1 and IF3. In this event, IF1 and IF3 as well as the mRNA are bound to the 30S ribosomal subunit (below).

Demonstration of the binding of an mRNA to a ribosomal subunit required isolation and separation of the 30S ribosomal subunit, an RNA fraction of the cell, and the purification of initiation factor proteins from the bacterial cells. This was followed by reconstitution (adding the separated fractions back together) in the correct order show that mRNA would only bind to the 30S subunit in the presence of the two specific initiation factor proteins.
206 Translation Initiation: mRNA Associates with 30S Ribsomal Subunit
Next, with the help of GTP and another initiation factor (IF2), the initiator fmettRNAf recognizes and binds to the AUG start codon found in all mRNAs. Some call the resulting structure (shown below) the Initiation Complex, which includes the 30S ribosomal subunit, Ifs 1, 2 and 3, and the fmet-tRNAf.
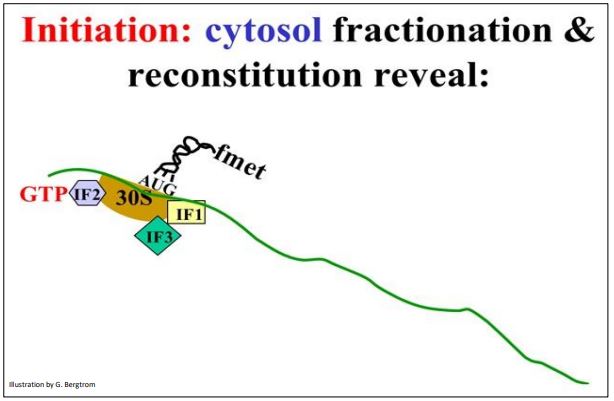
207 Initiation Complex Formation
In the last step of initiation, the large ribosomal subunit binds to this complex. IFs 1, 2 and 3 disassociate from the ribosome and the initiator fmet-tRNAfmet ends up in the P site of the ribosome.
Some prefer to call the structure formed at this point the initiation complex (below).
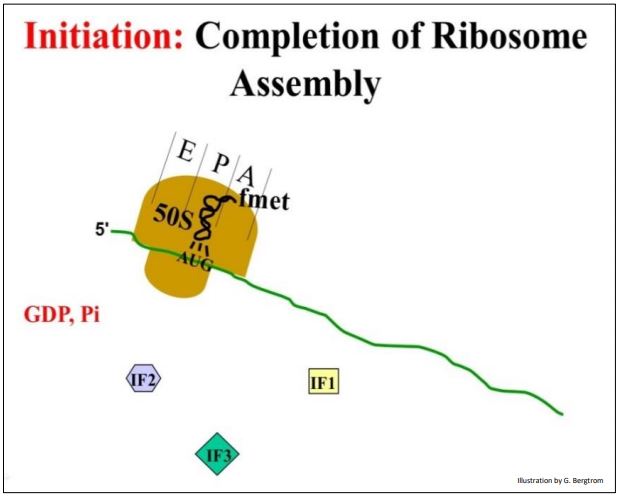
208 Adding the Large Ribosomal Subunit
Initiation can happen multiple times on a single mRNA, forming the polyribosome, or polysome described in Chapter 1. Each of the complexes formed above will engage in the elongation of a polypeptide described next.
3. Elongation
Elongation is a sequence of protein factor-mediated condensation reactions and ribosome movements along an mRNA. As you will see, polypeptide elongation requires a considerable input of free energy.
a) Elongation 1
The first step in elongation is the entry of the next aminoacyl-tRNA (aa2- tRNAaa2), which requires the free energy of GTP hydrolysis. The energy is supplied by the hydrolysis of GTP bound elongation factor 2 (EF2-GTP). The aa2-tRNAaa2 enters the ribosome based on codon-anticodon interaction at the A site as shown below.
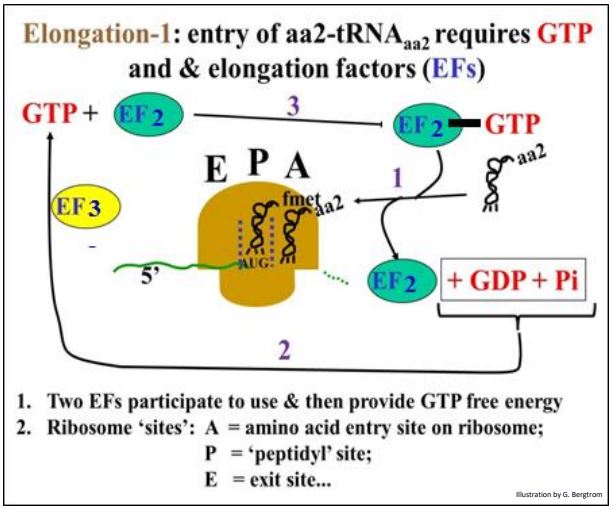
The GDP dissociates from EF2 as aa2-tRNAaa2 binds the anticodon in the A site. To keep elongation moving along, elongation factor (EF3) rephosphorylates the GDP to GTP, which can re-associate with free EF2.
209 Elongation: Elongation Factors and GTP
b) Elongation 2
Peptidyl transferase, a ribozyme component of the ribosome itself, links the incoming amino acid to a growing chain in a condensation reaction.
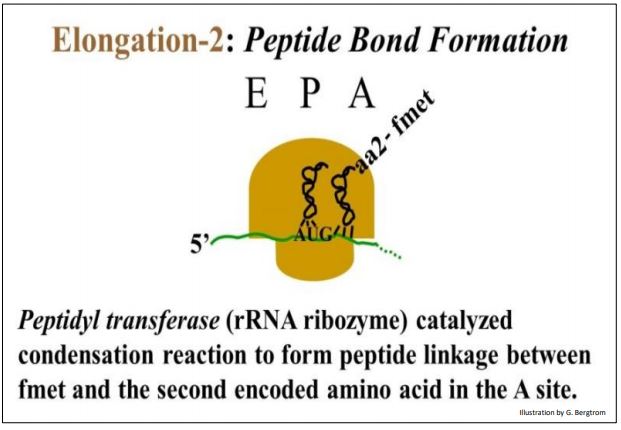
In this reaction, the fmet is transferred from the initiator tRNAf in the P site to aa2-tRNAaa2 in the A site, forming a peptide linkage with aa2.
210 Elongation: A Ribozyme Catalyzes Peptide Linkage Formation
c) Elongation 3
Translocase catalyzes GTP hydrolysis as the ribosome moves (translocates) along the mRNA. After translocation, the next mRNA codon shows up in the A site of the ribosome and the first tRNA (in this example, tRNAf) ends up on the E site of the ribosome.
The movement of the ribosome along the mRNA is illustrated below.
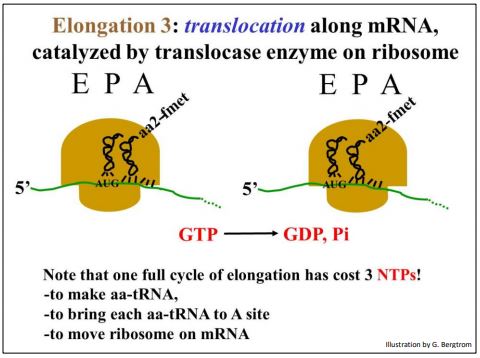
The tRNAf, no longer attached to an amino acid, will exit the E site as the next (3rd) aa-tRNA enters the empty A site, based on a specific codon-anticodon interaction (assisted by elongation factors and powered by GTP hydrolysis) to begin another cycle of elongation. Note that in each cycle of elongation, an ATP is consumed to attach each amino acid to its tRNA, and two GTPs are hydrolyzed in the cycle itself. In other words, at the cost of three NTPs, protein synthesis is the most expensive polymer synthesis reaction in cells!
211 Elongation: Translocase Moves Ribosomes along mRNA
212 Adding the Third Amino Acid
213 Big Translation Energy Costs
As polypeptides elongate, they eventually emerge from a groove in the large ribosomal subunit. As noted, a formylase enzyme in E. coli cytoplasm removes the formyl group from the exposed initiation fmet from all growing polypeptides. While about 40% of E. coli polypeptides still begin with methionine, specific proteases catalyze the hydrolytic removal of the amino-terminal methionine (and sometimes even more amino acids) from the other 60% of polypeptides. The removal of the formyl group and one or more N-terminal amino acids from new polypeptides are examples of post-translational processing.
214 The Fates of fMet and Met: Cases of Post-Translational Processing
4. Termination
Translation of an mRNA by a ribosome ends when translocation exposes one of the three stop codons in the A site of the ribosome. Stop codons are not situated some distance from the 3’ end of an mRNA. The region between a stop codon to the end of the mRNA is called the 3’ untranslated region of the messenger RNA (3’UTR).
Since there is no aminoacyl-tRNA with an anticodon to the stop codons (UAA, UAG or UGA), the ribosome actually stalls and the translation slow-down is just long enough for a protein termination factor to enter the A site. This interaction causes release of the new polypeptide and the disassembly of the ribosomal subunits from the mRNA. The process requires energy from yet another GTP hydrolysis. After dissociation, ribosomal subunits can be reassembled with an mRNA for another round of protein synthesis. Translation termination is illustrated below.
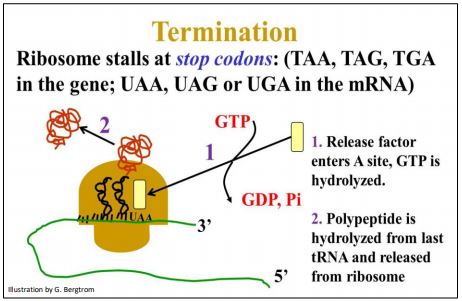
215 Translation Termination
We have seen some examples of post-translational processing (removal of formyl groups in E. coli, removal of the N-terminal methionine from most polypeptides, etc.) Most proteins, especially in eukaryotes, undergo one or more additional steps of posttranslational processing before becoming biologically active. We will see examples in upcoming chapters.
Let’s conclude this chapter with a “we thought we knew everything” moment! A recent study reports that ribosomes can sometimes re-initiate translation in the 3’ UTR of an mRNA using AUG codons upstream of the normal start codon of the mRNA. There is evidence that the resulting short polypeptides may be functional! Click here to read more: here.


