Serious efforts to understand how proteins are encoded began after Watson and Crick used the experimental evidence of Maurice Wilkins and Rosalind Franklin (among others) to determine the structure of DNA. Most hypotheses about the genetic code assumed that DNA (i.e., genes) and polypeptides were colinear.
A. Colinearity
For genes and proteins, colinearity just means that the length of a DNA sequence in a gene is proportional to the length of the polypeptide encoded by the gene. The gene mapping experiments in E. coli already discussed certainly supported this hypothesis.
The concept of colinearity is illustrated below.
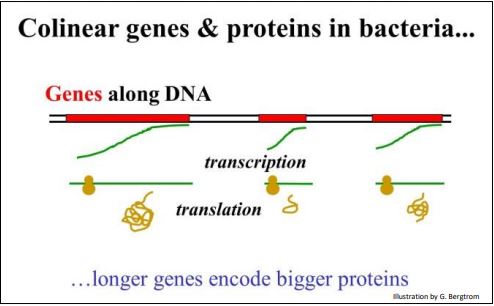
If the genetic code is collinear with the polypeptides it encodes, then a one-base codon obviously does not work because such a code would only account for four amino acids. A two-base genetic code also doesn’t work because it could only account for 16 (4 2 ) of the twenty amino acids found in proteins. However, threenucleotide codons could code for a maximum of 43 or 64 amino acids, more than enough to encode the 20 amino acids. And of course, a 4-base code also works; it satisfies the expectation that genes and proteins are collinear, with the’ advantage’ that there would be 256 possible codons to choose from (i.e., 44 possibilities).
B. How is the Genetic Code 'Read' to Account for All of an Organisms' Gene?
George Gamow (a Russian Physicist working at George Washington University) was the first to propose triplet codons to encode the twenty amino acids, the simplest hypothesis to account for the colinearity of gene and protein, and for encoding 20 amino acids. One concern that was raised was whether there is enough DNA in an organism’s genome to fit the all codons it needs to make all of its proteins? Assuming genomes did not have a lot of extra DNA laying around, how might genetic information be compressed into short DNA sequences in a way that is consistent with the colinearity of gene and polypeptide. One idea assumed 44 meaningless and 20 meaningful 3-base codons (one for each amino acid) and 44 meaningless codons, and that the meaningful codons in a gene (i.e., an mRNA) would be read and translated in an overlapping manner.
A code where codons overlap by one base is shown below.
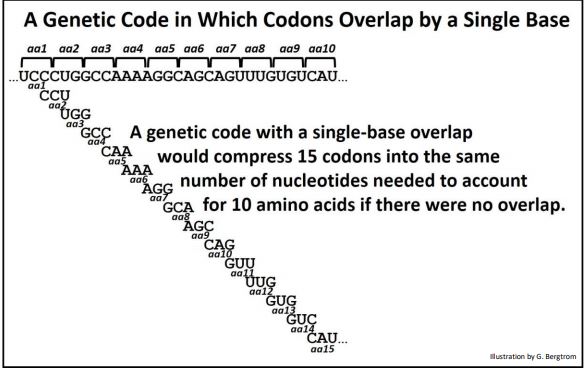
You can figure out how compressed a gene could get with codons that overlapped by two bases. However, as attractive as an overlapping codon hypothesis was in achieving genomic economies, it sank of its own weight almost as soon as it was floated! If you look carefully at the example above, you can see that each succeeding amino acid would have to start with a specific base. A look back at the table of 64 triplet codons quickly shows that only one of 16 amino acids, those that begin with a C can follow the first one in the illustration. Based on amino acid sequences accumulating in the literature, virtually any amino acid could follow another in a polypeptide. Therefore, overlapping genetic codes are untenable. The genetic code must be non-overlapping!
Sidney Brenner and Frances Crick performed elegant experiments that directly demonstrated the non-overlapping genetic code. They showed that bacteria with a single base deletion in the coding region of a gene failed to make the expected protein. Likewise, deleting two bases from the gene. On the other hand, bacteria containing a mutant version of the gene in which three bases were deleted were able to make the protein. The protein it made was slightly less active than bacteria with genes with no deletions.
The next issue was whether there were only 20 meaningful codons and 44 meaningless ones. If only 20 triplets actually encoded amino acids, how would the translation machinery recognize the correct 20 codons to translate? What would prevent the translational machinery from ‘reading the wrong’ triplets, i.e., reading an mRNA out of phase? If for example, if the translation machinery began reading an MRNA from the second or third bases of a codon, it would likely encounter a meaningless 3-base sequence in short order.
One speculation was that the code was punctuated. That is, perhaps there were the chemical equivalent of commas between the meaningful triplets. The commas would be of course, additional nucleotides. In such a punctuated code, the translation machinery would recognize the ‘commas’ and would not translate any meaningless 3- base triplet, avoiding out-of-phase translation attempts. Of course, a code with nucleotide ‘commas’ would increase the amount of DNA needed to specify a polypeptide by a third!
Then, Crick proposed the Commaless Genetic Code. He divided the 64 triplets into 20 meaningful codons that encoded the amino acids, and 44 meaningless ones that did not. The result was such that when the 20 meaningful codons are placed in any order, any of the triplets read in overlap would be among the 44 meaningless codons. In fact, he could arrange several different sets of 20 and 44 triplets with this property! Crick had cleverly demonstrated how to read the triplets in correct sequence without nucleotide ‘commas’.
202 Speculations About a Triplet Code
As we know now, the genetic code is indeed ‘commaless’… but not in the sense that Crick had envisioned. What’s more, Thanks to the experiments described next, we know that ribosomes read the correct codons in the right order because they know exactly where to start!
C. Breaking the Genetic Code
When the genetic code was actually broken, it was found that 61 of the codons specify amino acids and therefore, that the code is degenerate. Breaking the code began when Marshall Nirenberg and Heinrich J. Matthaei decoded the first triplet. They fractionated E. coli and identified which fractions had to be added back together in order to get polypeptide synthesis in a test tube (in vitro translation).
The cell fractionation is summarized below.
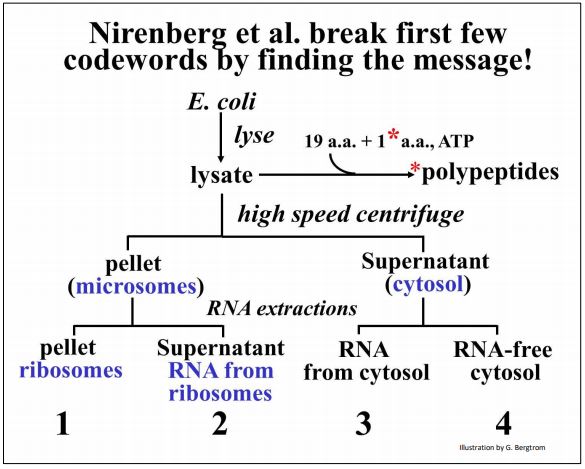
Check out the original work in the classic paper by Nirenberg MW and Matthaei JH [(1961) The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribo-nucleotides. Proc. Natl. Acad. Sci. USA 47:1588-1602]. The various cell fractions isolated by this protocol were added back together along with amino acids (one of which was radioactive) and ATP as an energy source. After a short incubation, Nirenberg and his coworkers looked for the presence of high molecular weight radioactive proteins as evidence of cell-free protein synthesis.
They found that all four final sub-fractions (1-4 above) must be added together to make radioactive proteins in the test tube. One of the essential cell fractions consisted of RNA that had been gently extracted from ribosome (fraction 2 in the illustration). Reasoning that this RNA might be mRNA, they substituted a synthetic poly(U) preparation for this fraction in their cell-free protein synthesizing mix, expecting poly(U) to encode a simple repeating amino acid.
They set up 20 reaction tubes, with a different amino acid in each…, and made only poly-phenylalanine. The experiment is illustrated below.
So, the triplet codon UUU means phenylalanine. Other polynucleotides were synthesized by G. Khorana, and in quick succession, poly(A) and poly(C) were shown to make poly-lysine and poly-proline in this experimental protocol. Thus AAA and CCC must encode lysine and proline respectively. With a bit more difficulty and ingenuity, poly di- and tri-nucleotides were also used in the cell free system to decipher several additional codons.
203 Deciphering the First Codon
M. W. Nirenberg, H. G. Khorana and R. W. Holley shared the 1968 Nobel Prize in Physiology or Medicine for their contributions to our understanding of protein synthesis. Deciphering the rest of the genetic code was based on Crick’s realization that chemically, amino acids have no attraction for either DNA or RNA (or triplets thereof). Instead, he predicted the existence of an adaptor molecule that would contain nucleic acid and amino acid information on the same molecule. Today we recognize this molecule as tRNA, the genetic decoding device.
Nirenberg and Philip Leder designed the experiment that pretty much broke the rest of the genetic code. They did this by adding individual amino acids to separate test tubes containing tRNAs, in effect causing the synthesis of specific aminoacyl-tRNAs.
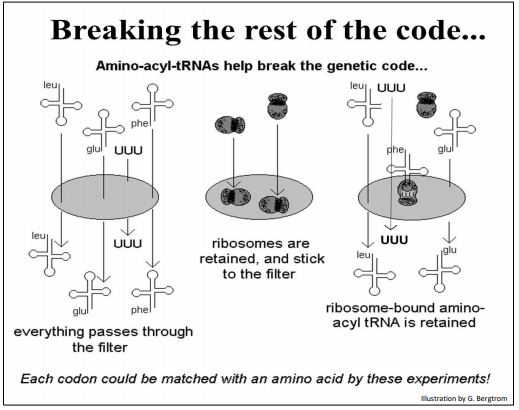
They then mixed their amino acid-bound tRNAs with isolated ribosomes and synthetic triplets. Since they had already shown that synthetic three-nucleotide fragments would bind to ribosomes, they hypothesized that triplet-bound ribosomes would in turn, bind appropriate amino acid-bound tRNAs. The experiment is shown below.

Various combinations of tRNA, ribosomes and aminoacyl-tRNAs were placed over a filter. Nirenberg and Leder knew that aminoacyl-tRNAs alone passed through the filter and that ribosomes did not. They predicted then, that triplets would associate with the ribosomes, and further, that this complex would bind the tRNA with the amino acid encoded by the bound triplet. This 3-part complex would also be retained by the filter, allowing the identification of the amino acid retained on the filter, and therefore the triplet code-word that had enabled binding the amino acid to the ribosome.
204 Deciphering all 64 Triplet Codons
After the code was largely deciphered, Robert Holley actually sequenced a yeast tRNA, and from regions of internal complementarity, predicted the folded structure of the tRNA. This first successful sequencing of a nucleic acid was possible because the tRNA was short, and contained several modified bases that facilitated the sequencing chemistry. Holley found the amino acid alanine at one end of the tRNA and he found one of the anticodons for an alanine codon roughly in the middle of the tRNA sequence. Holley predicted that this (and other) tRNAs would fold and assume a stem-loop, or cloverleaf structure with a central anticodon loop. The illustration below shows this structure for a phenylalanine tRNA along with subsequent computer-generated structures (below right) showing a now familiar “L”-shaped molecule with an amino acid attachment site at the 3’-end at the top of the molecule, and the anticodon loop at the other, bottom ‘end’

205 tRNA Structure and Base Modifications
After a brief overview of translation, we’ll break translation down into its 3 steps and see how aminoacyl-tRNAs function in the initiation and elongation steps of translation, as well as the special role of an initiator tRNA.


