Progress through the cell cycle is regulated. The cycle can be controlled or put on ‘pause’ at any one of several phase transitions. Such checkpoints monitor whether the cell is on track to complete a successful cell division event. Superimposed on these controls are signals that promote cell differentiation. Embryonic cells differentiate as the embryo develops. Even after terminal differentiation of cells that form all adult tissues and organs, adult stem cells will divide and differentiate to replace worn out cells. Once differentiated, cells are typically signaled in G1 to enter G0 and stop cycling. In some circumstances cells in G0 are recruited to resume cycling. However, if this occurs to by mistake, the cells may be transformed to cancer cells. Here we consider how the normal transition between phases of the cell cycle is controlled.
Growing, dividing cells monitor their progress through the phases. Cells produce internal chemical signals that tell them when it’s time to begin replication or mitosis, or even when to enter into G0 when they reach their terminally differentiated state. The experiment that first demonstrated a chemical regulator of the cell cycle involved fusing very large frog’s eggs! The experiment is described below.

The hypothesis tested here was that frog oocyte cytoplasm from germinal vesicle stage oocytes (i.e., in mid-meiosis) contains a chemical that caused the cell to lose its nuclear membrane, condense its chromatin into chromosomes and enter meiosis. Cytoplasm was withdrawn from one of these mid-meiotic oocytes with a fine hypodermic needle, and then injected into a pre-meiotic oocyte. The mid-meiotic oocyte cytoplasm induced premature meiosis in the immature oocyte. A maturation promoting factor (MPF) could be isolated from the mid-meiotic cells and injected into pre-meiotic cells; it caused them to enter meiosis. MPF turns out to be a protein kinase made up of two polypeptide subunits as shown below.
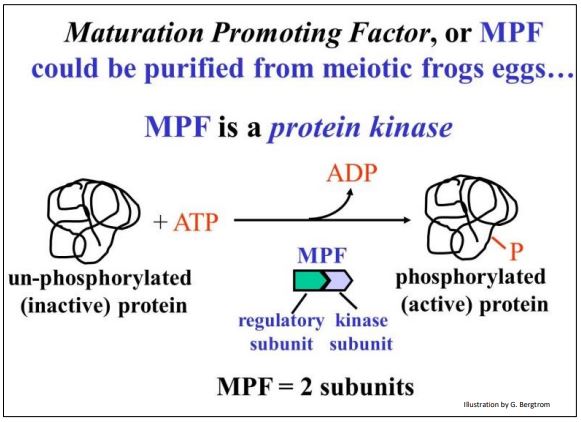
MPF was then also shown to stimulate somatic cells in G2 to enter premature mitosis. So conveniently, MPF can also be Mitosis Promoting Factor! Hereafter we will discuss the effects of MPF as being equivalent in mitosis and meiosis. When active, MPF targets many cellular proteins.
342 Discovery of MPF Kinase and Its Role in Meiosis and Mitosis
Assays of MPF activity as well as the actual levels of the two subunits over time during the cell cycle are graphed below.
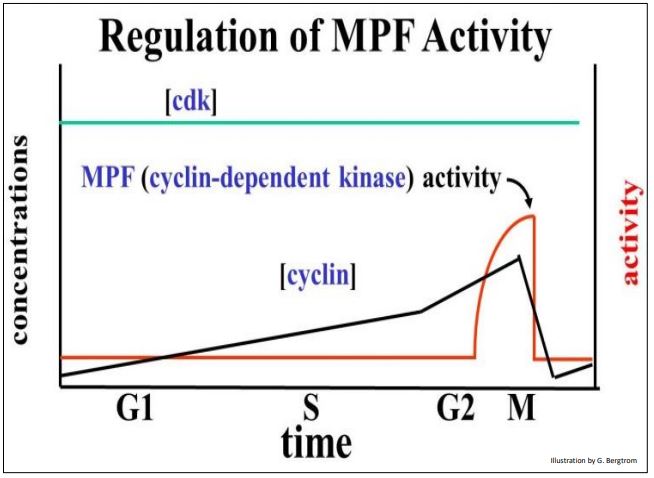
One subunit of MPF is cyclin, a regulatory polypeptide. The other subunit, cyclin-dependent kinase (cdk), contains the kinase enzyme active site. Both subunits must be bound to make an active kinase. Cyclin was so-named because its levels rise gradually after cytokinesis, peak at the next mitosis, and then fall. Levels of the cdk subunit do not change significantly during the life of the cell. Because the kinase activity of MPF requires cyclin, it tracks the rise in cyclin near the end of the G2, and its fall after mitosis. Cyclin begins to accumulate in G1, rising gradually and binding to more and more cdk subunits. MPF reaches a threshold concentration in G2 that triggers entry into mitosis. For their discovery of these central molecules Leland H. Hartwell, R. Timothy Hunt, and Paul M. Nurse won the 2001 Nobel Prize in Physiology or Medicine.
B. Other Cyclins, CDKs and Cell Cycle Checkpoints
Other chemical signals accumulate at different points in the cell cycle. For example, when cells in S are fused with cells in G1, the G1 cells begin synthesizing DNA (visualized as 3H-thymine incorporation). An experiment showing control of progress to different phases of the cell cycle is illustrated below.
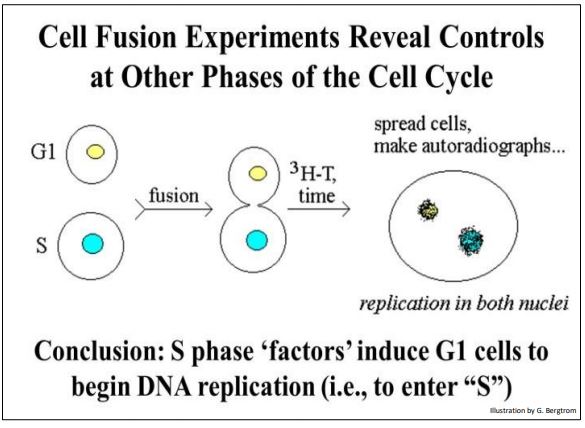
An S phase factor could be isolated from the S phase cells. This factor also turns out to be a two-subunit protein kinase, albeit a different one from MPF. Just as MPF signals cells in G2 to begin mitosis, the S phase kinase signals cells in G1 to enter the S phase of the cell cycle. MPF and the S phase kinase govern activities at two of several cell cycle checkpoints. In each case, the activity of the kinases is governed by prior progress through the cell cycle. In other words, if the cell is not ready to begin mitosis, active MPF production is delayed until it is. Likewise, the S phase kinase will not be activated until the cell is ready to begin DNA synthesis.
343 Cell Cycle Control at Check Points and the Go "Phase"
The sequence of signals that control progress through the cell cycle is probably more intricate and extensive than we currently know, but the best-described checkpoints are in G1, G2 and M (below).
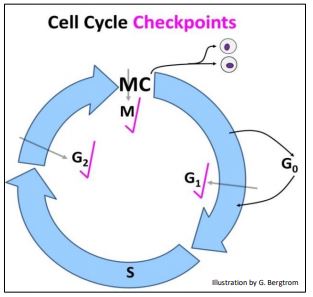
We generally envision checkpoints as monitoring and blocking progress until essential events of a current phase of the cell cycle phase are completed. These kinases are part of molecular sensing mechanisms that act by phosphorylating cytoplasmic and/or nuclear proteins required by upcoming phases of the cycle. Let’s take a closer look at some events that are monitored at these checkpoints in more detail.
1. The G1 Checkpoint
The G1 checkpoint controls the transition from the G1 to the S phase of the cell cycle. If actively dividing cells (e.g., stem cells) in G1 fail to complete their preparation for replication, the S-phase kinase won’t be produced and the cells won’t proceed the S phase until the preparatory biochemistry catches up with the rest of the cycle. To enter S, a cell must be ready to make proteins of replication, like DNA polymerases, helicases, and primases among others. Only when these molecules have accumulated to (or become active at) appropriate levels, is it “safe” to enter S and begin replicating DNA. This description of G1 checkpoint activity is consistent with the idea that all checkpoints delay cycling until a prior phase is complete.
What about cells that are fully differentiated? Such terminally differentiated cells stop producing the active G1 checkpoint kinase and stop dividing. These cells are arrested in G0 (see below). As an interesting side-note, recall that somatic cells are diploid and germ cells (sperm, egg) are haploid. So, are cells in G2 that have already doubled their DNA ‘tetraploid’, however briefly? Whether or not we can call G2 cells tetraploid (officially, probably not), it is clear that G1 cells and G0 cells are diploid!
2. The G2 Checkpoint
Passage through the G2 checkpoint is only possible if DNA made in the prior S phase is not damaged. Or if it was, that the damage has been (or can be) repaired (review the proofreading functions of DNA polymerase and the various DNA repair pathways). Cells that do successfully complete replication and pass the G2 checkpoint must prepare to make the proteins necessary for the upcoming mitotic phase. These include nuclear proteins necessary to condense chromatin into chromosomes, tubulins for making microtubules, etc. Only when levels of these and other required proteins reach a threshold can the cell begin mitosis. Consider the following two tasks required of the G2 checkpoint (in fact, any checkpoint):
- sensing whether prior phase activities have been successfully completed.
- delaying transition to the next phase if those activities are unfinished.
But what if sensing is imperfect and a checkpoint is leaky? A recent study suggests that either the G2 checkpoint is leaky, or at least, that incomplete activities in the S phase are tolerated, and that some DNA repair is not resolved until mitosis is underway in M! Check it out at DNA repair and replication during mitosis.
3. M Checkpoint
The M checkpoint is monitored by the original MPF phosphorylation of proteins that: (a) bind to chromatin causing it to condense and form chromatids, (b) lead to the breakdown of the nuclear envelope, and (c) enable spindle fiber formation,. In addition, tension in the spindle apparatus at metaphase tugs at the kinetochores holding the duplicated chromatids together. When this tension reaches a threshold, MPF peaks and an activated separase enzyme causes the chromatids to separate at their centromeres. Beginning in anaphase, tension in the spindle apparatus draws the new chromosomes to opposite poles of the cell. Near the end of mitosis and cytokinesis, proteins phosphorylated by MPF initiate the breakdown of cyclin in the cell. Passing the M checkpoint means that the cell will complete mitosis and cytokinesis, and that each daughter cell will enter a new G1 phase.
Dividing yeast cells only seem to have the three checkpoints discussed here. More complex eukaryotes use more cyclins and cdks to control the cell cycle at additional checkpoints. Different cyclins show cyclic patterns of synthesis, while cdks remain at constant levels throughout the cell cycle (as in MPF). Different gene families encode evolutionarily conserved cdks or cyclins. But each cyclin/cdk pair has been coopted in evolution to monitor different cell cycle events and to catalyze phosphorylation of phase-specific proteins. To learn more, see Elledge SJ (1996) Cell Cycle Checkpoints: Preventing an Identity Crisis. Science 274:1664-1672.
344 Cyclin/cdk Checkpoint for Cell Cycle Phases
C. The G0 State
This is not really a phase of the cell cycle, since cells in G0 have reached a terminally differentiated state and have stopped dividing. In development, terminally differentiated cells in tissues and organs no longer divide. Nevertheless, most cells have finite half-lives (recall our red blood cells that must be replaced every 60 days or so). Because cells in many tissues are in G0 and can’t divide, they must be replaced by stem cells, which can divide and differentiate. Some cells live so long in G0 that they are nearly never replaced (muscle cells, neurons). Other cells live short lives in G0 (e.g., stem cells, some embryonic cells). For example, a lymphocyte is a differentiated immune system white blood cell type. However, exposure of lymphocytes to foreign chemicals or pathogens activates mitogens that cause them to re-enter the cell cycle from G0. The newly divided cells then make the antibodies that neutralize the chemicals and fight off the pathogens. The retinoblastoma (Rb) protein is an example of a mitogen. Like other mitogens, the Rb protein is a transcription factor that turns on genes that lead to cell proliferation. What if cells continue cycling when they aren’t supposed to? Or, what if they are inappropriately signaled to exit Go? Such cells are in trouble! Having escaped normal controls on cell division, they can become a focal point of cancer cell growth. You can guess from its name that the retinoblastoma gene was discovered as a mutation that causes retinal cancer. For more about the normal function of the Rb protein and its interaction with a G1 cdk, check out the link below.
What if cells continue cycling when they aren’t supposed to? Or, what if they are inappropriately signaled to exit G0? Such cells are in trouble! Having escaped normal controls on cell division, they can become a focal point of cancer cell growth. You can guess from its name that the retinoblastoma gene was discovered as a mutation that causes retinal cancer. For more about the normal function of the Rb protein and its interaction with a G1 cdk, check out the link below.
345 Rb Gene Encodes Transcription Factor Regulatory Subunit


