10.3: Prokaryotic Gene Regulation
- Page ID
- 35721
What you’ll learn to do: Understand the basic steps in gene regulation in prokaryotic cells
The DNA of prokaryotes is organized into a circular chromosome supercoiled in the nucleoid region of the cell cytoplasm. Proteins that are needed for a specific function are encoded together in blocks called operons. For example, all of the genes needed to use lactose as an energy source are coded next to each other in the lactose (or lac) operon.
In prokaryotic cells, there are three types of regulatory molecules that can affect the expression of operons: repressors, activators, and inducers. Repressors are proteins that suppress transcription of a gene in response to an external stimulus, whereas activators are proteins that increase the transcription of a gene in response to an external stimulus. Finally, inducers are small molecules that either activate or repress transcription depending on the needs of the cell and the availability of substrate.
Understand the basic steps in gene regulation in prokaryotic cells
In bacteria and archaea, structural proteins with related functions—such as the genes that encode the enzymes that catalyze the many steps in a single biochemical pathway—are usually encoded together within the genome in a block called an operon and are transcribed together under the control of a single promoter. This forms a polycistronic transcript (Figure 1). The promoter then has simultaneous control over the regulation of the transcription of these structural genes because they will either all be needed at the same time, or none will be needed.

French scientists François Jacob (1920–2013) and Jacques Monod at the Pasteur Institute were the first to show the organization of bacterial genes into operons, through their studies on the lac operon of E. coli. They found that in E. coli, all of the structural genes that encode enzymes needed to use lactose as an energy source lie next to each other in the lactose (or lac) operon under the control of a single promoter, the lac promoter. For this work, they won the Nobel Prize in Physiology or Medicine in 1965.
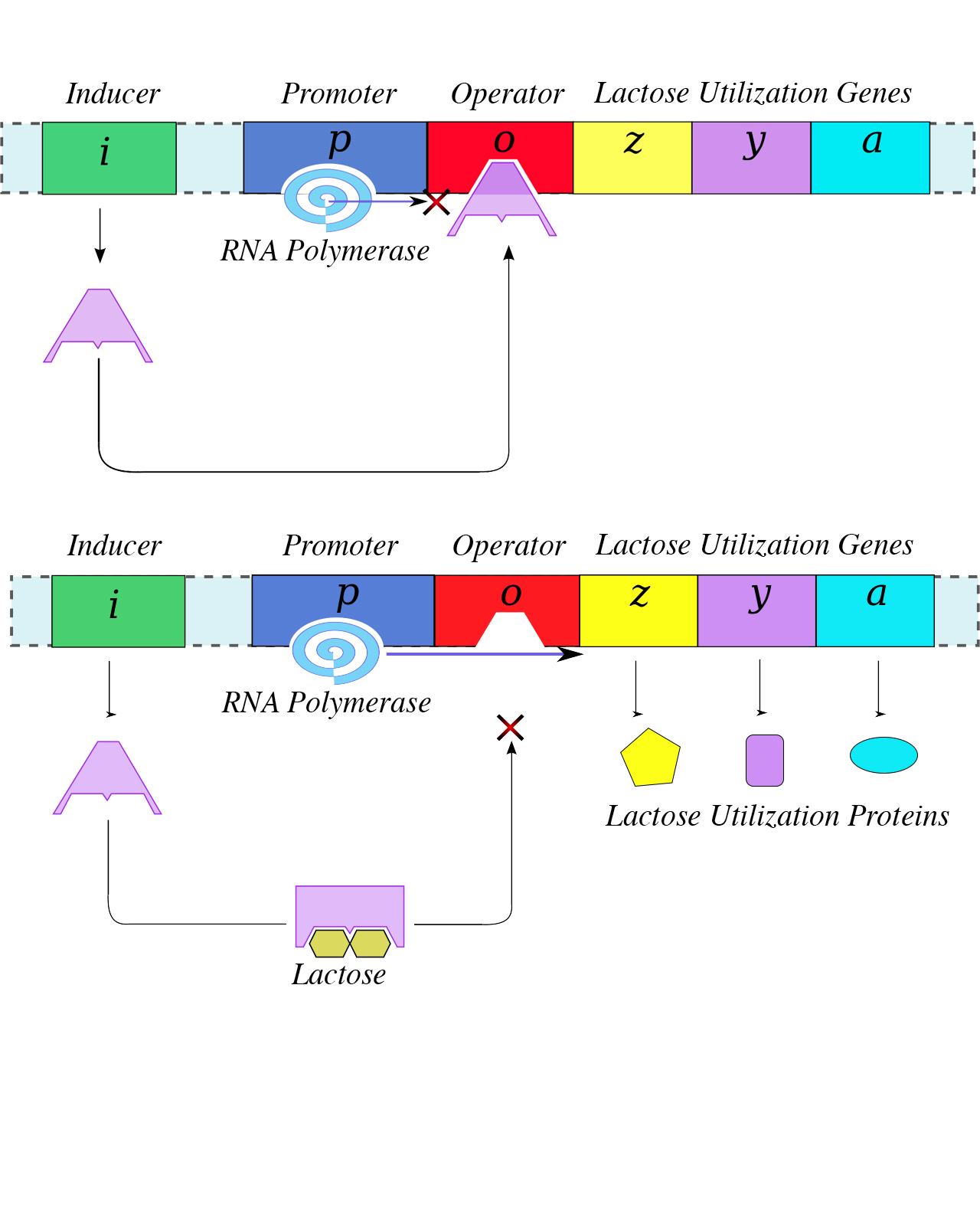 Each operon includes DNA sequences that influence its own transcription; these are located in a region called the regulatory region. The regulatory region includes the promoter and the region surrounding the promoter, to which transcription factors, proteins encoded by regulatory genes, can bind. Transcription factors influence the binding of RNA polymerase to the promoter and allow its progression to transcribe structural genes. A repressor is a transcription factor that suppresses transcription of a gene in response to an external stimulus by binding to a DNA sequence within the regulatory region called the operator, which is located between the RNA polymerase binding site of the promoter and the transcriptional start site of the first structural gene. Repressor binding physically blocks RNA polymerase from transcribing structural genes. Conversely, an activator is a transcription factor that increases the transcription of a gene in response to an external stimulus by facilitating RNA polymerase binding to the promoter. An inducer, a third type of regulatory molecule, is a small molecule that either activates or represses transcription by interacting with a repressor or an activator.
Each operon includes DNA sequences that influence its own transcription; these are located in a region called the regulatory region. The regulatory region includes the promoter and the region surrounding the promoter, to which transcription factors, proteins encoded by regulatory genes, can bind. Transcription factors influence the binding of RNA polymerase to the promoter and allow its progression to transcribe structural genes. A repressor is a transcription factor that suppresses transcription of a gene in response to an external stimulus by binding to a DNA sequence within the regulatory region called the operator, which is located between the RNA polymerase binding site of the promoter and the transcriptional start site of the first structural gene. Repressor binding physically blocks RNA polymerase from transcribing structural genes. Conversely, an activator is a transcription factor that increases the transcription of a gene in response to an external stimulus by facilitating RNA polymerase binding to the promoter. An inducer, a third type of regulatory molecule, is a small molecule that either activates or represses transcription by interacting with a repressor or an activator.
Other genes in prokaryotic cells are needed all the time. These gene products will be constitutively expressed, or turned on continually. Most consitutively expressed genes are “housekeeping” genes responsible for overall maintenance of a cell.
What are the parts in the DNA sequence of an operon?
- Show Answer
-
An operon is composed of a promoter, an operator, and the structural genes. They must occur in that order.
What types of regulatory molecules are there?
- Show Answer
-
There are three types of regulatory molecules: repressors, activators, and inducers.
Check Your Understanding
Answer the question(s) below to see how well you understand the topics covered in the previous section. This short quiz does not count toward your grade in the class, and you can retake it an unlimited number of times.
Use this quiz to check your understanding and decide whether to (1) study the previous section further or (2) move on to the next section.
Contributors and Attributions
- Introduction to Prokaryotic Gene Regulation. Provided by: Lumen Learning. License: CC BY: Attribution
- Lac Operon - a derivative from the Wikipedia image. Authored by: Stephen Snyder. Located at: https://commons.wikimedia.org/wiki/File:Operon_presence_lactose_numeros.svg. License: CC BY-SA: Attribution-ShareAlike
- Biology. Provided by: OpenStax CNX. Located at: http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8. License: CC BY: Attribution. License Terms: Download for free at http://cnx.org/contents/185cbf87-c72...f21b5eabd@10.8

