10.4: Eukaryotic Gene Regulation
- Page ID
- 35722
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)What you’ll learn to do: Discuss different components and types of epigenetic gene regulation
Your amazing body contains hundreds of different cell types, from immune cells to skin cells to neurons. Almost all of your cells contain the same set of DNA instructions: so why do they look so different, and do such different jobs? The answer: different gene regulation!
Gene regulation is how a cell controls which genes, out of the many genes in its genome, are “turned on” (expressed). Thanks to gene regulation, each cell type in your body has a different set of active genes – despite the fact that almost all the cells of your body contain the exact same DNA. These different patterns of gene expression cause your various cell types to have different sets of proteins, making each cell type uniquely specialized to do its job.
Eukaryotic gene expression is more complex than prokaryotic gene expression because the processes of transcription and translation are physically separated. Unlike prokaryotic cells, eukaryotic cells can regulate gene expression at many different levels. Eukaryotic gene expression begins with control of access to the DNA. This form of regulation, called epigenetic regulation, occurs even before transcription is initiated.
- Explain the process of epigenetic regulation
- Discuss the role of transcription factors in gene regulation
- Understand RNA splicing and explain its role in regulating gene expression
- Describe the importance of RNA stability in gene regulation
Eukaryotic Epigenetic Gene Regulation
The human genome encodes over 20,000 genes; each of the 23 pairs of human chromosomes encodes thousands of genes. The DNA in the nucleus is precisely wound, folded, and compacted into chromosomes so that it will fit into the nucleus. It is also organized so that specific segments can be accessed as needed by a specific cell type.
The first level of organization, or packing, is the winding of DNA strands around histone proteins. Histones package and order DNA into structural units called nucleosome complexes, which can control the access of proteins to the DNA regions (Figure 1a). Under the electron microscope, this winding of DNA around histone proteins to form nucleosomes looks like small beads on a string (Figure 1b). These beads (histone proteins) can move along the string (DNA) and change the structure of the molecule.

If DNA encoding a specific gene is to be transcribed into RNA, the nucleosomes surrounding that region of DNA can slide down the DNA to open that specific chromosomal region and allow for the transcriptional machinery (RNA polymerase) to initiate transcription (Figure 2). Nucleosomes can move to open the chromosome structure to expose a segment of DNA, but do so in a very controlled manner.

In females, one of the two X chromosomes is inactivated during embryonic development because of epigenetic changes to the chromatin. What impact do you think these changes would have on nucleosome packing?
- Show Answer
-
The nucleosomes would pack more tightly together.
This type of gene regulation is called epigenetic regulation. Epigenetic means “around genetics.” The changes that occur to the histone proteins and DNA do not alter the nucleotide sequence and are not permanent. Instead, these changes are temporary (although they often persist through multiple rounds of cell division) and alter the chromosomal structure (open or closed) as needed. A gene can be turned on or off depending upon the location and modifications to the histone proteins and DNA.
View this video that describes how epigenetic regulation controls gene expression.
In eukaryotic cells, the first stage of gene expression control occurs at the epigenetic level. Epigenetic mechanisms control access to the chromosomal region to allow genes to be turned on or off. These mechanisms control how DNA is packed into the nucleus by regulating how tightly the DNA is wound around histone proteins. The addition or removal of chemical modifications (or flags) to histone proteins or DNA signals to the cell to open or close a chromosomal region. Therefore, eukaryotic cells can control whether a gene is expressed by controlling accessibility to transcription factors and the binding of RNA polymerase to initiate transcription.
Eukaryotic Transcription Gene Regulation
Like prokaryotic cells, the transcription of genes in eukaryotes requires the actions of an RNA polymerase to bind to a sequence upstream of a gene to initiate transcription. However, unlike prokaryotic cells, the eukaryotic RNA polymerase requires other proteins, or transcription factors, to facilitate transcription initiation. Transcription factors are proteins that bind to the promoter sequence and other regulatory sequences to control the transcription of the target gene. RNA polymerase by itself cannot initiate transcription in eukaryotic cells. Transcription factors must bind to the promoter region first and recruit RNA polymerase to the site for transcription to be established.
View the process of transcription—the making of RNA from a DNA template:
The Promoter and the Transcription Machinery

Genes are organized to make the control of gene expression easier. The promoter region is immediately upstream of the coding sequence. The purpose of the promoter is to bind transcription factors that control the initiation of transcription.
Enhancers and Transcription
In some eukaryotic genes, there are regions that help increase or enhance transcription. These regions, called enhancers, are not necessarily close to the genes they enhance. They can be located upstream of a gene, within the coding region of the gene, downstream of a gene, or may be thousands of nucleotides away. Enhancer regions are binding sequences, or sites, for transcription factors. When a DNA-bending protein binds, the shape of the DNA changes (Figure 3). This shape change allows for the interaction of the activators bound to the enhancers with the transcription factors bound to the promoter region and the RNA polymerase.
Turning Genes Off: Transcriptional Repressors
Like prokaryotic cells, eukaryotic cells also have mechanisms to prevent transcription. Transcriptional repressors can bind to promoter or enhancer regions and block transcription. Like the transcriptional activators, repressors respond to external stimuli to prevent the binding of activating transcription factors.
To start transcription, transcription factors, must first bind to the promoter and recruit RNA polymerase to that location. In addition to promoter sequences, enhancer regions help augment transcription. Enhancers can be upstream, downstream, within a gene itself, or on other chromosomes. Transcription factors bind to enhancer regions to increase or prevent transcription.
The binding of ________ is required for transcription to start.
- a protein
- DNA polymerase
- RNA polymerase
- a transcription factor
- Show Answer
-
Answer c. The binding of RNA polymerase is required for transcription to start.
What will result from the binding of a transcription factor to an enhancer region?
- decreased transcription of an adjacent gene
- increased transcription of a distant gene
- alteration of the translation of an adjacent gene
- initiation of the recruitment of RNA polymerase
- Show Answer
-
Answer b. Increased transcription of a distant gene will result from the binding of a transcription factor to an enhancer region.
A mutation within the promoter region can alter transcription of a gene. Describe how this can happen.
- Show Answer
-
A mutation in the promoter region can change the binding site for a transcription factor that normally binds to increase transcription. The mutation could either decrease the ability of the transcription factor to bind, thereby decreasing transcription, or it can increase the ability of the transcription factor to bind, thus increasing transcription.
What could happen if a cell had too much of an activating transcription factor present?
- Show Answer
-
If too much of an activating transcription factor were present, then transcription would be increased in the cell. This could lead to dramatic alterations in cell function.
Post-Transcriptional Control of Gene Expression
RNA is transcribed, but must be processed into a mature form before translation can begin. This processing after an RNA molecule has been transcribed, but before it is translated into a protein, is called post-transcriptional modification. As with the epigenetic and transcriptional stages of processing, this post-transcriptional step can also be regulated to control gene expression in the cell. If the RNA is not processed, shuttled, or translated, then no protein will be synthesized.
RNA splicing, the first stage of post-transcriptional control
In eukaryotic cells, the RNA transcript often contains regions, called introns, that are removed prior to translation. The regions of RNA that code for protein are called exons (Figure 4). After an RNA molecule has been transcribed, but prior to its departure from the nucleus to be translated, the RNA is processed and the introns are removed by splicing.
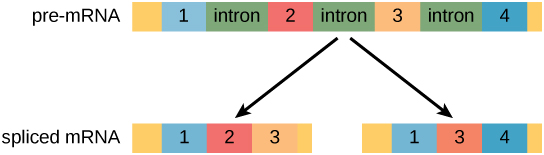
Alternative RNA Splicing
Alternative RNA splicing is a mechanism that allows different protein products to be produced from one gene when different combinations of introns, and sometimes exons, are removed from the transcript (Figure 5).

This alternative splicing can be haphazard, but more often it is controlled and acts as a mechanism of gene regulation.
Visualize how mRNA splicing happens by watching the process in action in this video:
Control of RNA Stability
Before the mRNA leaves the nucleus, it is given two protective “caps” that prevent the end of the strand from degrading during its journey. The 5′ cap, which is placed on the 5′ end of the mRNA and poly-A tail, which is attached to the 3′ end. Once the RNA is transported to the cytoplasm, the length of time that the RNA resides there can be controlled. Each RNA molecule has a defined lifespan and decays at a specific rate. This rate of decay is referred to as the RNA stability. If the RNA is stable, it will be detected for longer periods of time in the cytoplasm.
RNA Stability and microRNAs
The microRNAs, or miRNAs, are short single-stranded RNA molecules that are only 21–24 nucleotides in length. Like transcription factors and RBPs, mature miRNAs recognize a specific sequence and bind to the RNA to degrade the target mRNA. They rapidly destroy the RNA molecule.
Post-transcriptional regulation can occur at any stage after transcription, including RNA splicing, nuclear shuttling, and RNA stability. Once RNA is transcribed, it must be processed to create a mature RNA that is ready to be translated. This involves the removal of introns that do not code for protein. Spliceosomes bind to the signals that mark the exon/intron border to remove the introns and ligate the exons together. Once this occurs, the RNA is mature and can be translated. RNA is created and spliced in the nucleus, but needs to be transported to the cytoplasm to be translated. RNA is transported to the cytoplasm through the nuclear pore complex. Once the RNA is in the cytoplasm, the length of time it resides there before being degraded, called RNA stability, can also be altered to control the overall amount of protein that is synthesized. RNA stability is controlled by microRNAs (miRNAs). These miRNAs bind to the 5′ CAP or the 3′ Tail of the RNA to decrease RNA stability and promote decay.
Which of the following are involved in post-transcriptional control?
- control of RNA splicing
- control of RNA shuttling
- control of RNA stability
- all of the above
- Show Answer
-
Answer d. All of the above (control of RNA splicing, RNA shuttling, and RNA stability) are involved in post-transcriptional control.
Binding of a miRNAs will ________ the stability of the RNA molecule.
- increase
- decrease
- neither increase nor decrease
- either increase or decrease
- Show Answer
-
Answer b. Binding of a miRNAs will decrease the stability of the RNA molecule.
Check Your Understanding
Answer the question(s) below to see how well you understand the topics covered in the previous section. This short quiz does not count toward your grade in the class, and you can retake it an unlimited number of times.
Use this quiz to check your understanding and decide whether to (1) study the previous section further or (2) move on to the next section.
Contributors and Attributions
- Introduction to Eukaryotic Gene Regulation. Provided by: Lumen Learning. License: CC BY: Attribution
- Biology. Provided by: OpenStax CNX. Located at: http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8. License: CC BY: Attribution. License Terms: Download for free at http://cnx.org/contents/185cbf87-c72...f21b5eabd@10.8
- Overview: Eukaryotic gene regulation. Provided by: Khan Academy. Located at: https://www.khanacademy.org/science/biology/gene-regulation/gene-regulation-in-eukaryotes/a/overview-of-eukaryotic-gene-regulation. License: CC BY-NC-SA: Attribution-NonCommercial-ShareAlike

