Chapter 10: Microbial Metabolism II
- Page ID
- 146532
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
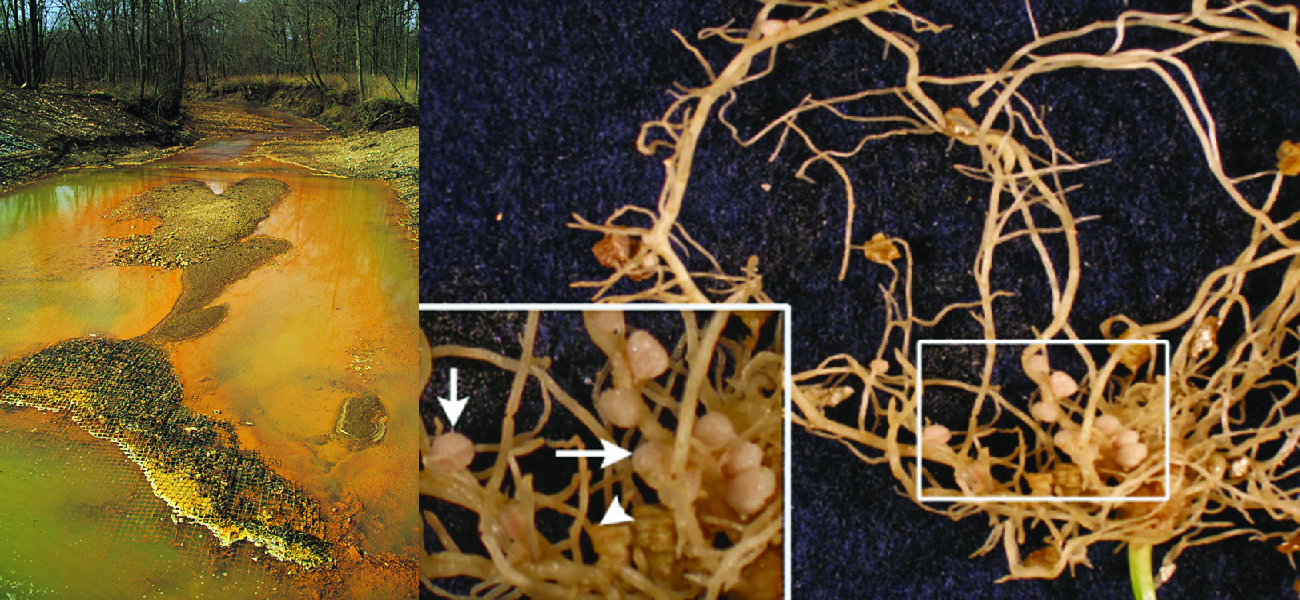
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Throughout earth’s history, microbial metabolism has been a driving force behind the development and maintenance of the planet’s biosphere. Eukaryotic organisms such as plants and animals typically depend on organic molecules for energy, growth, and reproduction. Prokaryotes, on the other hand, can metabolize a wide range of organic as well as inorganic matter, from complex organic molecules like cellulose to inorganic molecules and ions such as atmospheric nitrogen (N2), molecular hydrogen (H2), sulfide (S2−), manganese (II) ions (Mn2+), ferrous iron (Fe2+), and ferric iron (Fe3+), to name a few. By metabolizing such substances, microbes chemically convert them to other forms. In some cases, microbial metabolism produces chemicals that can be harmful to other organisms; in others, it produces substances that are essential to the metabolism and survival of other life forms (Figure \(\PageIndex{1}\)).
- Section 10.1: Cellular Respiration
- Cellular respiration begins when electrons are transferred from NADH and FADH₂—through a series of chemical reactions to a final inorganic electron acceptor (either oxygen in aerobic respiration or non-oxygen inorganic molecules in anaerobic respiration). These electron transfers take place on the inner part of the cell membrane of prokaryotic cells or in specialized protein complexes in the inner membrane of the mitochondria of eukaryotic cells.
- Section 10.2: Fermentation
- Fermentation uses an organic molecule as a final electron acceptor to regenerate NAD⁺ from NADH so that glycolysis can continue. Fermentation does not involve an electron transport system, and no ATP is made by the fermentation process directly. Fermenters make very little ATP—only two ATP molecules per glucose molecule during glycolysis. Microbial fermentation processes have been used for the production of foods and pharmaceuticals, and for the identification of microbes.
- Section 10.3: Catabolism of Lipids and Proteins
- Collectively, microbes have the ability to degrade a wide variety of carbon sources besides carbohydrates, including lipids and proteins. The catabolic pathways for all of these molecules eventually connect into glycolysis and the Krebs cycle. Several types of lipids can be microbially degraded. Triglycerides are degraded by extracellular lipases, releasing fatty acids from the glycerol backbone. Phospholipids are degraded by phospholipases, releasing fatty acids and phosphorylated head groups.
- Section 10.4: Photosynthesis and the Importance of Light
- Heterotrophic organisms ranging from E. coli to humans rely on the chemical energy found mainly in carbohydrate molecules. Many of these carbohydrates are produced by photosynthesis, the biochemical process by which phototrophic organisms convert solar energy (sunlight) into chemical energy. Although photosynthesis is most commonly associated with plants, microbial photosynthesis is also a significant supplier of chemical energy, fueling many diverse ecosystems.
- Section 10.5: Biogeochemical Cycles
- Energy flows directionally through ecosystems, entering as sunlight for phototrophs or as inorganic molecules for chemoautotrophs. The six most common elements associated with organic molecules—carbon, hydrogen, nitrogen, oxygen, phosphorus, and sulfur—take a variety of chemical forms and may exist for long periods in the atmosphere, on land, in water, or beneath earth’s surface.
Thumbnail: The Krebs cycle, also known as the citric acid cycle, is summarized here. Note incoming two-carbon acetyl results in the main outputs per turn of two CO2, three NADH, one FADH2, and one ATP (or GTP) molecules made by substrate-level phosphorylation. Two turns of the Krebs cycle are required to process all of the carbon from one glucose molecule. (CC BY 4.0; OpenStax)


