1.33: Bacterial Transformation
- Page ID
- 79457
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Describe and explain Griffith's experiment originating bacterial transformation.
- Define and properly use the following terms: transformation, recombinant DNA, transgenic, competent cells, biotechnology, vector, genetic engineering, plasmid, horizontal gene transfer, selectable marker, GFP.
- Tell at least two applications for bacterial transformation.
- Explain how bacterial transformation works.
- Successfully conduct a bacterial transformation.
- Interpret results from bacterial transformation.
The Origins of Bacterial Transformation
British bacteriologist Frederick Griffith reported the first demonstration of bacterial transformation—a process in which external DNA is taken up by a cell, thereby changing its morphology and physiology. Griffith conducted his experiments with Streptococcus pneumoniae, a bacterium that causes pneumonia. Griffith worked with two strains of this bacterium called rough (R) and smooth (S). The two cell types were called “rough” (R) and “smooth” (S) after the appearance of their colonies grown on a nutrient agar plate.
The R strain is non-pathogenic (does not cause disease). The S strain is pathogenic (disease-causing), and has a capsule outside its cell wall. The capsule allows the cell to escape the immune responses of the host mouse.
When Griffith injected the living S strain into mice, they died from pneumonia. In contrast, when Griffith injected the live R strain into mice, they survived. In another experiment, when he injected mice with the heat-killed S strain, they also survived. This experiment showed that the capsule alone was not the cause of death. In a third set of experiments, a mixture of live R strain and heat-killed S strain were injected into mice, and—to his surprise—the mice died. Upon isolating the live bacteria from the dead mouse, only the S strain of bacteria was recovered. When this isolated S strain was injected into fresh mice, the mice died. Griffith concluded that something had passed from the heat-killed S strain into the live R strain and transformed it into the pathogenic S strain. He called this the transforming principle. These experiments are now known as Griffith's transformation experiments.

Figure 1: Two strains of S. pneumoniae were used in Griffith’s transformation experiments. The R strain is non-pathogenic, whereas the S strain is pathogenic and causes death. When Griffith injected a mouse with the heat-killed S strain and a live R strain, the mouse died. The S strain was recovered from the dead mouse. Griffith concluded that something had passed from the heat-killed S strain to the R strain, transforming the R strain into the S strain in the process. Credit: Rao, A., Ryan, K. and Tag, A. Department of Biology, Texas A&M University.
What occurred in Griffith's experiments in 1928 could not be fully explained at that time. Scientists then were unaware of the role of DNA blueprints for a living thing. Further, with such limited knowledge about DNA and what it does, scientists then were also unaware that bacteria are capable of "picking up" DNA from other bacterial cells or picking up DNA from their environment.
In Griffith's experiment, DNA from the dead S strain were transferred into the live R strain. This DNA provided the R strain with instructions for building the S strain's capsule. As a result, the live R strain cells began to produce the S strain's capsule and thereby developed the ability to evade the host's immune system. The result is the R strain acted in the same way as the S strain, became pathogenic, and caused death in the mice. In essence, what happened in these circumstances is bacterial transformation. Bacterial transformation is a process where bacteria absorb DNA from their environment resulting in new characteristics in the bacteria. The new characteristics that transformed bacteria (also called transgenic bacteria) exhibit is dependent on what types of genes are present in the DNA that they took up from their environment.
transgenic: describes a living thing that has DNA introduced into it through artificial means; the DNA added to the transgenic organism originates from a different species
Why Transform Bacteria?
Using living systems to benefit humankind is called biotechnology. Technically speaking, the domestication of plants and animals through farming and breeding practices is a type of biotechnology, however in a contemporary sense, we associate biotechnology with the direct alteration of an organism’s genetics to achieve desirable traits through the process of genetic engineering.
Genetic engineering involves the use of recombinant DNA technology, the process by which a DNA sequence is manipulated in vitro, thus creating recombinant DNA molecules that have new combinations of genetic material. The recombinant DNA is then introduced into a host organism. If the DNA that is introduced comes from a different species, the host organism is considered to be transgenic.

Figure 2: Bacterial transformation commonly uses a plasmid to carry a gene of interest into a bacterial cell. During transformation, a bacterial cell takes up the plasmid. Afterward, that bacterial cell will contain the genes in that plasmid and can express those genes.
One example of a transgenic microorganism is the bacterial strain that produces human insulin. The insulin gene from humans was inserted into a plasmid. This recombinant DNA plasmid was then inserted into bacteria. As a result, these transgenic microbes have the DNA necessary to produce and secrete human insulin.
Many prokaryotes have the ability to acquire foreign DNA and incorporate functional genes into their own genome through “mating” with other cells (conjugation), viral infection (transduction), and taking up DNA from the environment (transformation). These mechanisms of DNA transfers are examples of horizontal gene transfer—the transfer of genetic material between cells of the same generation.
Beyond applications that generate useful products for humans, transformation is incredibly useful in studying genetics. Transformation has enabled scientists to expand understanding of the functions of specific genes and regulation of gene expression (which genes are turned on/off in what circumstances, and how those genes are regulated).
How does Bacterial Transformation Work?
Plasmids
A gene of interest (a segment of DNA that codes for a gene to be inserted into a living thing) are commonly inserted into plasmids. Plasmids are small pieces of typically circular, double-stranded DNA that replicate independently of the bacterial chromosome. In recombinant DNA technology, plasmids are often used as vectors - they are used for transferring DNA. Plasmids can be used to carry DNA fragments from one organism to another.
Plasmids used as vectors can be genetically engineered by researchers and scientific supply companies to have specialized properties. Some plasmid vectors contain genes that confer a selectable marker to help researchers separate bacteria carrying the plasmid from bacteria not carrying the plasmid. One of the most common selectable markers used is antibiotic resistance genes. For this, the plasmid is engineered with an antibiotic resistance gene so that all bacteria carrying the plasmid are resistant to that antibiotic. If the bacteria are then plated on petri plates containing the antibiotic in the medium, then only those bacteria that carry the plasmid with its antibiotic resistance gene will grow. Bacteria without the plasmid will be killed by the presence of the antibiotic in the medium since they lack the antibiotic resistance gene. The colonies that grow in the presence of the antibiotic can therefore be used in the ongoing research knowing that they carry the plasmid.

Figure 3: Diagram of an example of an artificially constructed plasmid vector commonly used for cloning foreign DNA. This plasmid's name is pUC19 and it is 2,686 base pairs long. The red and pink arrows represent the regions of DNA in this plasmid. The arrows indicate the directions in which the genes are transcribed. Note the ampicillin resistance gene (amp) encoded on the plasmid is used as the selectable marker. Plasmids are also engineered with a polylinker site, containing multiple unique restriction enzyme recognition sites (these recognition sites are locations where enzymes called restriction enzymes can be used to cut the DNA of the plasmid to insert a gene of interest into the plasmid). The polylinker site in this example is positioned within the lacZ gene. The lacZ gene can be used as a reporter gene in order to determine if the gene of interest was successfully inserted into the plasmid. For more about this, see this page about blue-white screening.
Bacterial Transformation
The most commonly used mechanism for introducing engineered plasmids into a bacterial cell is transformation, a process in which bacteria take up free DNA from their surroundings. In nature, free DNA typically comes from other lysed bacterial cells. In the laboratory, free DNA in the form of recombinant plasmids is introduced to the cell’s surroundings.

Figure 4: Bacterial transformation commonly uses a plasmid to carry a gene of interest into a bacterial cell. During transformation, a bacterial cell takes up the plasmid. Afterward, that bacterial cell will contain the genes in that plasmid and can express those genes.
Some bacteria, such as Bacillus spp., are naturally competent, meaning they are able to take up foreign DNA. However, not all bacteria are naturally competent. In most cases, bacteria must be made artificially competent in the laboratory by increasing the permeability of the cell membrane. This can be achieved through chemical treatments that neutralize charges on the cell membrane or by exposing the bacteria to an electric field that creates microscopic pores in the cell membrane. These methods yield chemically competent or electrocompetent bacteria, respectively.

Figure 5: Bacterial transformation process shown with the example of the insulin gene. Recombinant DNA technology is the artificial recombination of DNA from two organisms. In this example, the human insulin gene is inserted into a bacterial plasmid. This recombinant plasmid can then be used to transform bacteria, which gain the ability to produce the insulin protein.
GFP
GFP stands for green fluorescent protein. This protein is coded in a gene that originated in the jellyfish species Aequorea victoria. GFP is commonly used in genetics research to visualize expression of a gene. GFP glows fluorescent green under ultraviolet light.
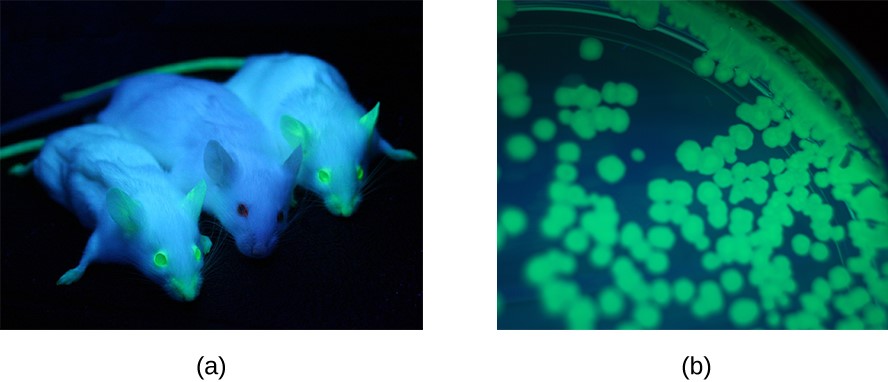
Figure 6: (a) The gene encoding green fluorescence protein is a commonly used reporter gene for monitoring gene expression patterns in organisms. Under ultraviolet light, GFP fluoresces. Here, two mice are expressing GFP, while the middle mouse is not. (b) GFP can be used as a reporter gene in bacteria as well. Here, a plate containing bacterial colonies expressing GFP are shown. (credit a: modification of work by Ingrid Moen, Charlotte Jevne, Jian Wang, Karl-Henning Kalland, Martha Chekenya, Lars A Akslen, Linda Sleire, Per Ø Enger, Rolf K Reed, Anne M Øyan, Linda EB Stuhr; credit b: modification of work by “2.5JIGEN.com”/Flickr)
Laboratory Instructions
Your instructor will provide instructions for the transformation protocol you will use in this laboratory.
Questions
- Define the following:
- transformation
- Recombinant DNA
- Genetic engineering
- Transgenic
- Biotechnology
- Vector
- Selectable marker
- Plasmid
- GFP
- Competent cells
- Cells are made competent using one of two different approaches. What are these?
- Explain how antibiotic resistance can be used as a selectable marker in genetic engineering.
- Why are selectable markers used in genetic engineering?
- Give at least two reasons it is useful to transform bacteria with certain genes of interest.
- What is the purpose of the heat-shock step during bacterial transformation?
- What would happen if you forgot to add the plasmid before the heat shock step? Would this change the results from this experiment? Explain.
- What was added into the medium in the petri plates to insure that only bacteria carrying the plasmid would grow?
Attributions
- Biology 2e by OpenStax is licensed under CC BY 4.0
- Microbiology by OpenStax is licensed under CC BY 4.0


