19.7A: Transcription
- Page ID
- 3434
- Define the following:
- gene
- transcription
- Briefly describethe function of the following in terms of transcription:
- mRNA
- 3' end
- 5' end
- RNA polymerase
- phosphodiester bond
- promoter
- leader sequence
- coding sequence
- transcription terminator
- codon
- Define the following in terms of transcription in eukaryotic cells:
- introns
- exons
- precurser mRNA
- cap
- poly-A tail
- mature mRNA
Transcription in Prokaryotic Cells
Description: Messenger RNA (mRNA) is synthesized by complementary base pairing of ribonucleotides with deoxyribonucleotides to match a portion of one strand of DNA called a gene. Although genes are present on both strands of DNA, only one strand is transcribed for any given gene. Following transcription, 30S and 50S ribosomal subunits attach to the mRNA and tRNA inserts the correct amino acids which are subsequently joined to form a polypeptide or a protein through a process called translation.
The enzyme RNA polymerase transcribes DNA. This enzyme initiates transcription, joins the RNA nucleotides together, and terminates transcription. To initiate transcription in bacteria, a variety of proteins called sigma factors bind to RNA polymerases. This complex can then bind to a specific sequence of usually about 40 deoxyribonucleotide bases called the promoter located along the DNA prior to the coding region of the gene. The promotor determines what region of the DNA and which strand of DNA will be transcribed into RNA.
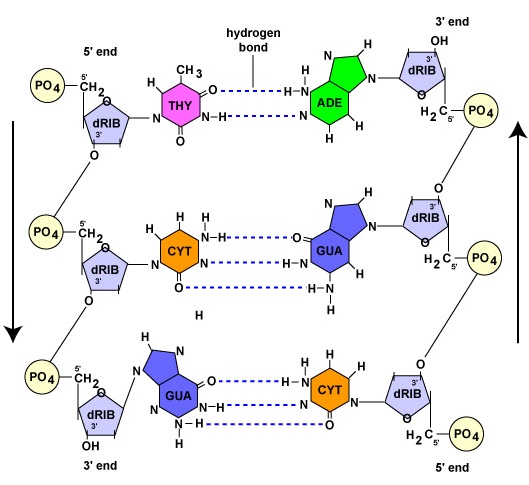
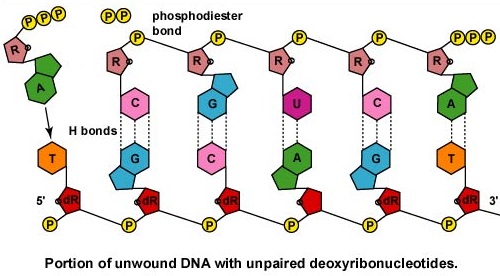
Like DNA polymerase, RNA polymerase can only synthesize nucleic acid in a 5' to 3' direction while "reading" a DNA template in the 3' to 5' direction. As mentioned earlier in this unit, the 3' end of a strand of nucleic acid has a hydroxyl (OH) group on the 3' carbon of the deoxyribose or ribose and is not linked to another nucleotide. The 5' end of that strand has a phosphate group attached to the 5' carbon of the sugar and is not linked to another nucleotide (Figure \(\PageIndex{1}\)).
Once the RNA polymerase/sigma factor complex recognizes the correct promoter, the sigma factor dissociate from the RNA polymerase and the enzyme begins to unwind the helix of the DNA creating a region of nonpaired deoxyribonucleotides that serve as a template for RNA synthesis (Figures 2 and 3).
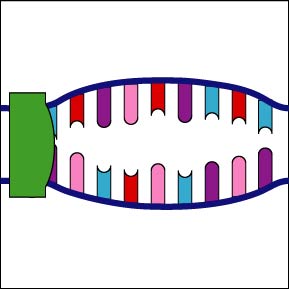
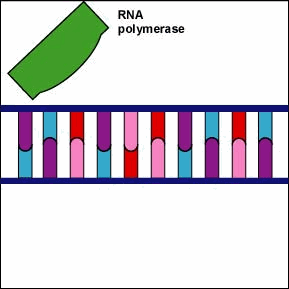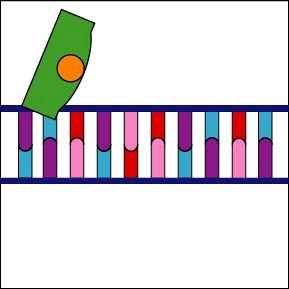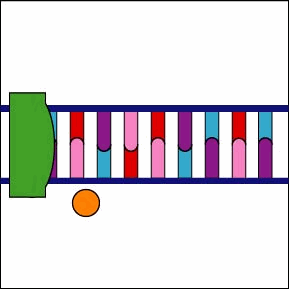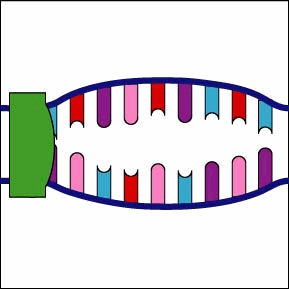
Once the RNA polymerase/sigma factor complex recognizes the correct promoter, the sigma factor dissociate from the RNA polymerase and the enzyme begins to unwind the helix of the DNA creating a region of nonpaired deoxyribonucleotides that serve as a template for RNA synthesis
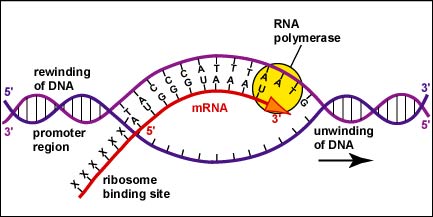
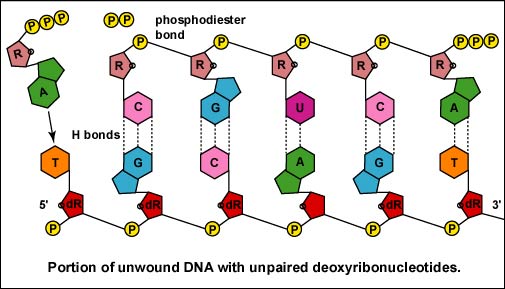
Once the actual transcription begins, ribonucleotides containing 3 phosphate groups hydrogen bond through the process of complementary base pairing with the exposed deoxyribonucleotides on the unwound strand that is to be transcribed (Figure \(\PageIndex{4}\)). The ribonucleotides are then covalently bonded together by phosphodiester bonds, the energy being supplied by the cleavage of two phosphate groups from the ribonucleotide triphosphate (Figure \(\PageIndex{5}\)). (The phosphodiester bond refers to the phosphate on the 5'C of the newly inserted nucleotide covalently bonding to the 3'C of the last ribonucleotide in the mRNA chain.) The mRNA polymerizes at a rate of about 30 nucleotides per second.
As the RNA polymerase moves down the DNA, the previous stretch of DNA again pairs with its complementary strand. This process continues until the RNA polymerase encounters a "stop" signal or transcription terminator at the end of the gene. This causes the completed mRNA to drop off the gene.
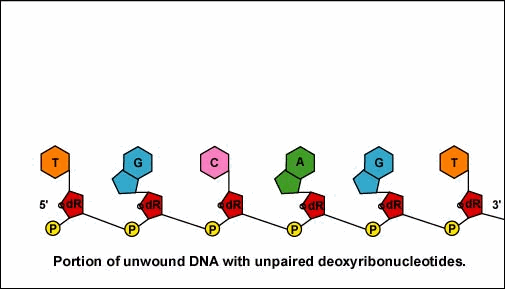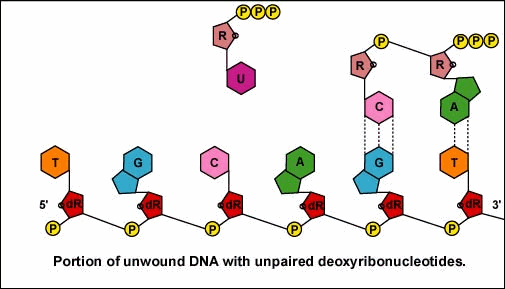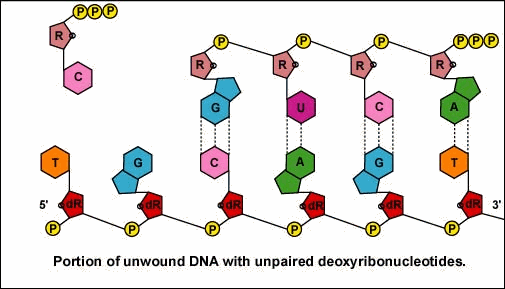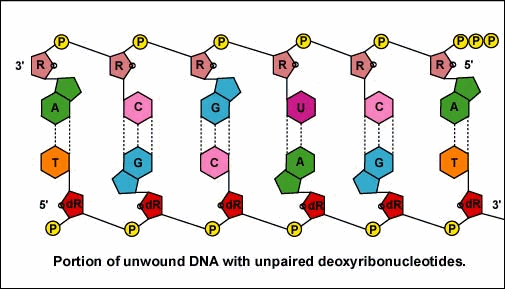
Once the RNA polymerase/sigma factor complex recognizes the correct promoter, the sigma factor dissociate from the RNA polymerase and the enzyme begins to unwind the helix of the DNA creating a region of nonpaired deoxyribonucleotides that serve as a template for RNA synthesis. Transcription is under control of the enzyme RNA polymerase which is not shown here.
Once the RNA polymerase moves beyond the promotor region, a new molecule of RNA polymerase can bind to the promotor and start a new round of transcription. In this way, a single gene can be transcribed multiple times.
|
YouTube movie illustrating complementary base pairing during transcription.
|
|
YouTube movie illustrating transcription and protein assembly.
|
|
YouTube movie illustrating transcription in bacteria
|
Transcription is summarized in Figs. 6 and 7.
There are 22 amino acids that can be encoded by the genetic information carried on mRNA. The mRNA molecule is divided up into codons. A codon is a series of three consecutive mRNA bases coding for one specific amino acid. The various codons and the amino acids for which they code are shown in Table \(\PageIndex{16}\).8.1. There are 64 codons. One codon, AUG, also serves as a start codon to initiate translation, and three codons, UAG, UAA, and UGA, function as stop or nonsense codons to terminate translation. (Alternative start codons are different from the standard AUG codon and are found occasionally in both prokaryotes and eukaryotes.)
|
U
|
C
|
A
|
G
|
||
|---|---|---|---|---|---|
|
U
|
UUU = Phe UUC = Phe UUA = Leu UUG = Leu |
UCU = Ser UCC = Ser UCA = Ser UCG = Ser |
UAU = Tyr UAC = Tyr UAA = Stop UAG = Stop |
UGU = Cys UGC = Cys UGA = Stop UGG = Trp |
U C A G |
|
C
|
CUU = Leu CUC = Leu CUA = Leu CUG = Leu |
CCU = Pro CCC = Pro CCA = Pro CCG = Pro |
CAU = His CAC = His CAA = Gln CAG = Gln |
CGU = Arg CGC = Arg CGA = Arg CGG = Arg |
U C A G |
|
A
|
AUU = Ile AUC = Ile AUA = Ile AUG = Met |
ACU = Thr ACC = Thr ACA = Thr ACG = Thr |
AAU = Asn AAC = Asn AAA = Lys AAG = Lys |
AGU = Ser AGC = Ser AGA = Arg AGG = Arg |
U C A G |
|
G
|
GUU = Val CUC = Val GUA = Val GUG = Val |
GCU = Ala GCC = Ala GCA = Ala GCG = Ala |
GAU = Asp GAC = Asp GAA = Glu GAG = Glu |
GGU = Gly GGC = Gly GGA = Gly GGG = Gly |
U C A G |
| Phe = phenylalanine Leu = leucine Ile = isoleucine Met = methionine Val = valine |
Ser = serine Pro = proline Thr = threonine Ala = alanine Tyr = tyrosine |
His = histidine Gln = glutamine Asn = asparagine Lys = lysine Asp = aspartic acid |
Glu = glutamic acid Cys = cysteine Trp = tryptophan Arg = arginine Gly = glycine |
||
| AUG = start codon, UAA, UAG, and UGA = stop (nonsense) codons | |||||
In bacteria, a mRNA can be monocistronic or polycistronic. A monocistronic mRNA is a transcript of a single gene. A polycistronic mRNA carries a transcript of multiple genes, often involved in a single biochemical pathway. Groups of related genes that are transcribed together to form a polycistronic mRNA are known as operons. There are also specific genes along the DNA from which each of the different transfer RNAs (tRNAs) and the ribosomal RNAs (rRNAs) are transcribed.
Most mRNAs in prokaryotes have a half-life on the order of a few minutes. Molecules of rRNA and tRNA, on the other hand, are much more stabile. Because rRNA and tRNA are highly folded molecules, unlike mRNA, they are much more resistant to degradation by ribonucleases. Once transcribed, the mRNA can be translated into protein.
Transcription in Eukaryotic Cells
Transcription is more complex in eukaryotic cells than in those that are prokaryotic. Activator proteins bind to genes known as enhancers which help determine which genes are switched on and speed up transcription. Repressor proteins bind to genes called silencers which interfere with activator proteins and slow down transcription. Coactivators, adapter molecules which coordinate signals from activator and repressor proteins, relay this information to basal factors which then position RNA polymerase at the start of the coding region of the gene to begin transcription.
Once the actual transcription begins, ribonucleotides containing 3 phosphate groups form hydrogen bonds through the process of complementary base pairing with the exposed deoxyribonucleotides on the unwound strand that is to be transcribed. The ribonucleotides are then covalently bonded together by phosphodiester bonds, the energy being supplied by the cleavage of two phosphate groups from the ribonucleotide triphosphate (Figure \(\PageIndex{16}\).8B.5). (The phosphodiester bond refers to the phosphate on the 5'C of the newly inserted nucleotide covalently bonding to the 3'C of the last ribonucleotide in the mRNA chain.)
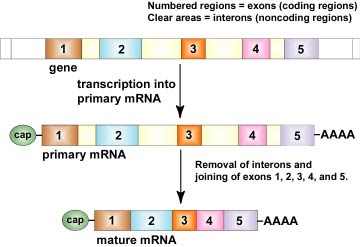
Unlike prokaryotes, most genes in higher eukaryotic cells contain large amounts - as much as 98% in the human genome - of regionscalled introns that are not part of the code for the final protein. These are interspersed among the coding regions or exons that actually code for the final protein.
RNA polymerase copies both the exons and the introns to form what is called precursor mRNA or pre-mRNA. Early in transcription, a cap in the form of an unusual nucleotide, 7-methylguanylate, is added to the 5' end of the pre-mRNA. This cap helps ribosomes attach for translation. As transcription is nearly completed, a series of 100-250 adenine ribonucleotides called a poly-A tail is added to the 3' end of the pre-mRNA. This poly-A tail is thought to help transport the mRNA out of the nucleus and may stabilize the mRNA against degradation in the cytoplasm. After transcription of the precursor mRNA, non-protein coding regions (introns) are excised and coding regions (exons) are joined together by complexes of ribonucleoproteins called spliceosomes to produce what is termed mature mRNA as shown in Figure \(\PageIndex{9}\). This process is called RNA processing.
|
YouTube movie illustrating complementary base pairing during transcription.
|
|
YouTube movie illustrating transcription and protein assembly.
|
|
YouTube movie illustrating RNA processing in eukaryotic cells #1.
|
|
YouTube movie illustrating RNA processing in eukaryotic cells #2.
|
The mature mRNA then passes through the pores in the nuclear membrane to be translated into protein by tRNA on eukaryotic 80S ribosomes (composed of 60S and 40S subunits) in a manner similar to prokaryotes.
The mRNA molecule is divided up into codons. A codon is a series of three consecutive mRNA bases coding for one specific amino acid. The various codons and the amino acids for which they code are shown in Figure \(\PageIndex{8}\). There are 64 codons. One codon, AUG, also serves as a start codon to initiate translation, and three codons, UAG, UAA, and UGA, function as stop or nonsense codons to terminate translation. (Alternative start codons are different from the standard AUG codon and are found occasionally in both prokaryotes and eukaryotes.)
In addition to the genes that are transcribed into mRNA to be translated into polypeptides and proteins, there are also specific genes in the DNA from which each of the different transfer RNAs (tRNAs) and the ribosomal RNAs (rRNAs) are transcribed.
Once transcribed, the mRNA can be translated into protein.
As mentioned above, introns make up the majority of DNA in higher eukaryotic cells and for decades was considered to be "junk DNA" accumulated over millions of years of evolution. Over recent years however, it has been discovered that much of this intergenic DNA, although it does not code for protein synthesis, is transcribed into functional molecules of RNA with names such as antisense RNA microRNA, and riboswitch RNA that play important roles in whether or not a protein is actually made.
Antisense RNA is RNA transcribed off of the strand of DNA complementary to the one being transcribed into mRNA. In other words, it is an RNA molecule complementary to a mRNA and as such may complementary base pair with the mRNA and prevents it from being translated into protein.
MicroRNA, often transcribed from intron DNA, folds over upon itself to resemble double-stranded RNA, a form of RNA produced by many viruses during their life cycle. Viral double-stranded RNA activates a host defense mechanism that degrades that viral RNA. The MicroRNA frequently binds to mRNA and tricks this defense mechanism into degrading that mRNA so it can not be translated into protein.
Riboswitch RNA, often transcribed from introns, exists in an inactive form until a specific target chemical binds. The binding of the target chemical turns the riboswitch RNA to an active form that can be translated into a specific protein.
Summary
- During protein synthesis, the order of nucleotide bases along a gene gets transcribed into a complementary strand of mRNA which is then translated by tRNA into the correct order of amino acids for that polypeptide or protein.
- The order of deoxyribonucleotide bases along the DNA determines the order of amino acids in the proteins, that is, its primary structure.
- Because certain amino acids can interact with other amino acids, the order of amino acids for each protein determines its final three-dimensional shape, which in turn determines the function of that protein.
- Messenger RNA (mRNA) is synthesized by complementary base pairing of ribonucleotides with deoxyribonucleotides to match a portion of one strand of DNA called a gene.
- Although genes are present on both strands of DNA, only one strand is transcribed for any given gene.
- The enzyme RNA polymerase transcribes DNA.
- To initiate transcription in bacteria, a variety of proteins called sigma factors bind to RNA polymerases. This complex can then bind to a specific DNA sequence called the promoter located along the DNA prior to the coding region of the gene. The promotor determines what region of the DNA and which strand of DNA will be transcribed into RNA.
- Like DNA polymerase, RNA polymerase can only synthesize nucleic acid in a 5' to 3' direction while "reading" a DNA template in the 3' to 5' direction.
- Once the RNA polymerase/sigma factor complex recognizes the correct promoter, the sigma factor dissociate from the RNA polymerase and the enzyme begins to unwind the helix of the DNA creating a region of nonpaired deoxyribonucleotides that serve as a template for RNA synthesis.
- During transcription, ribonucleotides hydrogen bond through the process of complementary base pairing with the exposed deoxyribonucleotides on the unwound strand that is to be transcribed. The ribonucleotides are then covalently bonded together by phosphodiester bonds.
- This process continues until the RNA polymerase encounters a "stop" signal or transcription terminator at the end of the gene.
- A single gene can be transcribed multiple times.
- The mRNA molecule is divided up into codons. A codon is a series of three consecutive mRNA bases coding for one specific amino acid.
- Three codons, UAG, UAA, and UGA, function as stop or nonsense codons to terminate translation.
- In bacteria, a mRNA can be monocistronic or polycistronic. A monocistronic mRNA is a transcript of a single gene; a polycistronic mRNA carries a transcript of multiple genes, often involved in a single biochemical pathway. Once transcribed, the mRNA can be translated into protein by tRNA on 70S ribosomes (composed of 50S and 30S subunits).
- Transcription is more complex in eukaryotic cells than in those that are prokaryotic. Activator proteins bind to genes known as enhancers which help determine which genes are switched on and speed up transcription. Repressor proteins bind to genes called silencers which interfere with activator proteins and slow down transcription. Coactivators, adapter molecules which coordinate signals from activator and repressor proteins, relay this information to basal factors which then position RNA polymerase at the start of the coding region of the gene to begin transcription.
- Most genes in higher eukaryotic cells contain regions called introns that are not part of the code for the final protein. These are interspersed among the coding regions or exons that actually code for the final protein.
- After transcription of the precursor mRNA, non-protein coding regions (introns) are excised and coding regions (exons) are joined together by complexes of ribonucleoproteins called spliceosomes to produce what is termed mature mRNA.
- The mature mRNA then passes through the pores in the nuclear membrane to be translated into protein by tRNA on 80S ribosomes (composed of 60S and 40S subunits) in a manner similar to prokaryotes.


