13.1B: Antibody Structure
- Page ID
- 3305
- Describe an antibody molecule.
- Draw the "stick figure" structure of IgG, indicating the Fab portion (variable region) and the Fc portion (constant region).
- State the functions of the Fab and the Fc portions of an antibody.
- State what is meant by the biological activity of an antibody.
- Compare the structure of IgM and secretory IgA with that of IgG.
In this section we will look at the structure of antibodies. There are five classes or isotypes of human antibodies :
- immunoglobulin G (IgG),
- immunoglobulin M (IgM),
- immunoglobulin A (IgA),
- immunoglobulin D (IgD), and
- immunoglobulin E (IgE).
The simplest antibodies, such as IgG, IgD, and IgE, are "Y"-shaped macromolecules called monomers. A monomer is composed of four glycoprotein chains: two identical heavy chains and two identical light chains. The two heavy chains have a high molecular weight that varies with the class of antibody. The light chains come in two varieties: kappa or lambda and have a lower molecular weight than the heavy chains. The four glycoprotein chains are connected to one another by disulfide (S-S) bonds and non-covalent bonds (Figure \(\PageIndex{1}\)).
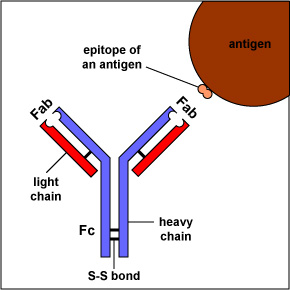
Additional S-S bonds fold the individual glycoprotein chains into a number of distinct globular domains (Figure \(\PageIndex{2}\)). The area where the top of the "Y" joins the bottom is called the hinge. This area is flexible to enable the antibody to bind to pairs of epitopes various distances apart on an antigen.
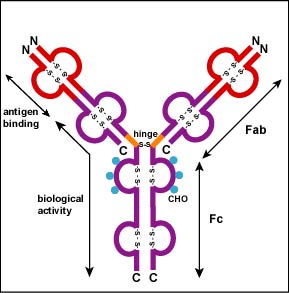
The two tips of the "Y" monomer are referred to as the antigen-binding fragments or Fab portions of the antibody (Figures 1-3). The first 110 amino acids or first domain of both the heavy and light chain of the Fab region of the antibody provide specificity for binding an epitope on an antigen. The amino acid sequence of this first domain of both the light chain and the heavy chain shows tremendous variation from antibody to antibody and constitutes the variable region (V region). This is because each B-lymphocyte, early in its development, becomes genetically programmed through a series of gene-splicing reactions to produce a Fab with a unique 3-dimensional shape capable of fitting some epitope with a corresponding shape.
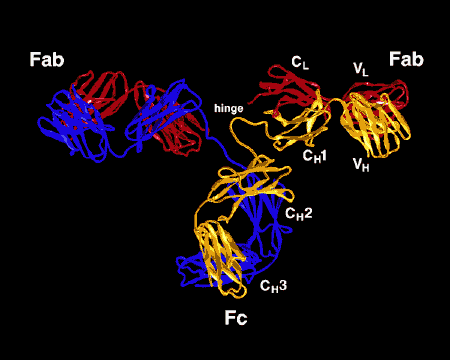
Figure \(\PageIndex{3}\): Ribbon Drawing of the Antibody Molecule IgG2a, A ribbon drawing of the first intact antibody molecule ever crystallized (IgG2a). The Fab portion of the antibody has specificity for binding an epitope of an antigen. The Fc portion directs the biological activity of the antibody.
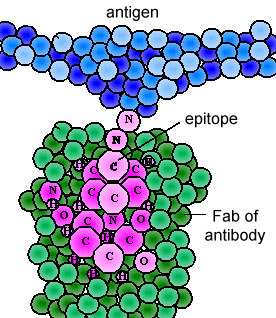
The various genes the cell splices together determine the order of amino acids of the Fab portion of both the light and heavy chain; the amino acid sequence determines the final 3-dimensional shape (Figure \(\PageIndex{4}\)). Therefore, different antibody molecules produced by different B-lymphocytes will have different orders of amino acids at the tips of the Fab to give them unique shapes for binding epitope. The antigen-binding site is large enough to hold an epitope of about 5-7 amino acids or 3-4 sugar residues. Epitopes bind to the Fab portion of the antibody by reversible, non-covalent bonds.
The bottom part of the "Y", the C terminal region of each glycoprotein chain, is called the Fc portion. The Fc portion, as well as one domain of both the heavy and light chain of the Fab region has a constant amino acid sequence and is referred to as the constant region (C region) of the antibody and defines the class and subclass of each antibody. The Fc portion is responsible for the biological activity of the antibody (Figures 1-3), however, the Fc portion only becomes biologically active after the Fab component has bound to its corresponding antigen. Depending on the class and subclass of antibody, biological activities of the Fc portion of antibodies include the ability to:
- Activate the classical complement pathway (IgG & IgM); see Figure \(\PageIndex{5}\).
- Activate the lectin complement pathway and the alternative complement pathway (IgA)
- Bind to receptors on phagocytes (IgG); see Figure \(\PageIndex{6}\).
- Bind to receptors on mast cells, basophils, and eosinophils (IgE); see Fig 7 and Figure \(\PageIndex{8}\).
- Bind to receptors on NK cells (IgG); see Figure \(\PageIndex{9}\).
- Determine the tissue distribution of the antibodies, that is, to what tissues types the antibody molecules are able to go.
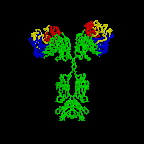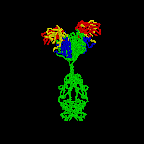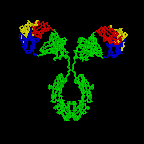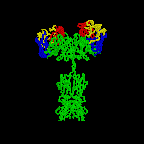
Individual "Y"-shaped antibody molecules are called monomers and can bind to two identical epitopes. Antibodies of the classes IgG, IgD, and IgE are monomers.
Two classes of antibodies are more complex. IgM (see Figure \(\PageIndex{10}\)) is a pentamer, consisting of 5 "Y"-like molecules connected at their Fc portions by a "J" or joining chain. Secretory IgA (see Figure \(\PageIndex{11}\)) is a dimer consisting of 2 "Y"-like molecules connected at their Fc portions by a "J" chain and stabilized to resist enzymatic digestion in body secretions by means of a secretory component.
Summary
- There are 5 classes or isotypes of human antibodies or immunoglobulins: IgG, IgM, IgA, IgD, and IgE.
- The simplest antibodies, such as IgG, IgD, and IgE, are "Y"-shaped macromolecules called monomers and are composed of four glycoprotein chains: two identical heavy chains and two identical light chains.
- The two tips of the "Y" monomer are referred to as the antigen-binding fragments or Fab portions of the antibody and these portions provide specificity for binding an epitope on an antigen.
- Early in its development, each B-lymphocyte becomes genetically programmed through a series of gene-splicing reactions to produce a Fab with a unique 3-dimensional shape capable of fitting some epitope with a corresponding shape.
- The Fc portion only becomes biologically active after the Fab component has bound to its corresponding antigen. Biological activities include activating the complement pathways, and binding to receptors on phagocytes and other defense cells to promote adaptive immunity.
- IgM is a pentamer, consisting of 5 monomers joined at their Fc portions.
- IgA is a dimer, consisting of 2 monomers joined at their Fc portions.
Questions
Study the material in this section and then write out the answers to these questions. Do not just click on the answers and write them out. This will not test your understanding of this tutorial.
- Describe an antibody molecule. (ans)
- Match the following:
_____ The region of the antibody that provide specificity for binding an epitope on an antigen. (ans)
_____ The region of the antibody that is responsible for the biological activity of the antibody. (ans)
_____ Composed of four glycoprotein chains. There are two identical heavy chains having a high molecular weight and two identical light chains. (ans)
_____ A pentamer, consisting of 5 "Y"-like molecules connected at their Fc portions by a "J" or joining chain. (ans)
_____ A dimer consisting of 2 "Y"-like molecules connected at their Fc portions by a "J" chain and stabilized to resist enzymatic digestion. (ans)
- IgM
- secretory IgA
- IgG
- Fab
- Fc
- Multiple Choice (ans)


